P61587
Gene name |
RND3 (ARHE, RHO8, RHOE) |
Protein name |
Rho-related GTP-binding protein RhoE |
Names |
Protein MemB, Rho family GTPase 3, Rho-related GTP-binding protein Rho8, Rnd3 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:390 |
EC number |
|
Protein Class |
|
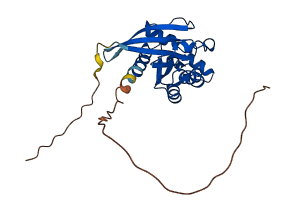
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

8 structures for P61587
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1M7B | X-ray | 200 A | A | 19-200 | PDB |
| 2V55 | X-ray | 370 A | B/D | 1-200 | PDB |
| 4BG6 | X-ray | 230 A | Q/R | 232-241 | PDB |
| 7OBC | X-ray | 190 A | B | 231-241 | PDB |
| 7OBD | X-ray | 200 A | B | 231-241 | PDB |
| 8B2K | X-ray | 140 A | B | 231-244 | PDB |
| 8BFC | X-ray | 140 A | B | 231-244 | PDB |
| AF-P61587-F1 | Predicted | AlphaFoldDB |
147 variants for P61587
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA348873000 rs1182539573 |
5 | R>K | No |
ClinGen TOPMed |
|
|
rs759966800 CA348872992 |
6 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs772557837 CA1902738 |
6 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs759966800 CA1902737 |
6 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA348872985 rs773861698 |
7 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1406655640 CA348872961 |
10 | L>F | No |
ClinGen TOPMed |
|
|
rs1467092859 CA348872960 |
11 | S>T | No |
ClinGen gnomAD |
|
|
rs1455403238 CA348872947 |
12 | S>R | No |
ClinGen gnomAD |
|
|
rs147918776 CA1902734 |
15 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 17 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA58436378 rs892934922 |
18 | P>S | No |
ClinGen Ensembl |
|
|
rs777448950 CA1902733 |
20 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769524149 CA348872882 |
21 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747961527 CA1902731 |
22 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs754971352 CA1902729 |
26 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA348872824 rs1337593899 |
30 | G>A | No |
ClinGen TOPMed |
|
| TCGA novel | 30 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1260221848 CA348872815 |
31 | D>E | No |
ClinGen gnomAD |
|
|
rs1348555628 CA348872822 |
31 | D>N | No |
ClinGen gnomAD |
|
|
CA348872802 rs1473735404 |
33 | Q>R | No |
ClinGen gnomAD |
|
|
CA1902727 rs779158844 |
34 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs754050173 CA1902725 |
36 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1054378795 CA58436377 |
38 | A>S | No |
ClinGen Ensembl |
|
|
rs753058821 CA1902722 |
39 | L>Q | No |
ClinGen ExAC gnomAD |
|
|
CA348872754 rs1279002171 |
41 | H>R | No |
ClinGen TOPMed |
|
|
rs760054624 CA1902720 |
47 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774796736 CA1902719 |
49 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs772797723 CA1902716 |
50 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348872691 rs1474730686 |
50 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA348872668 rs1251228402 |
51 | N>K | No |
ClinGen gnomAD |
|
|
rs759860450 CA1902699 |
53 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA348872649 rs1283966249 |
54 | P>L | No |
ClinGen gnomAD |
|
|
CA1902698 rs752108217 |
54 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs974493111 CA58436320 |
60 | Y>H | No |
ClinGen TOPMed |
|
|
rs766894438 CA1902697 |
61 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs766894438 CA58436318 |
61 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA58436319 rs1008533367 |
61 | T>P | No |
ClinGen Ensembl |
|
|
rs1186122682 CA348872594 |
63 | S>C | No |
ClinGen Ensembl |
|
|
CA58436317 rs1047476805 |
64 | F>L | No |
ClinGen Ensembl |
|
|
rs930357058 CA58436316 |
65 | E>K | No |
ClinGen Ensembl |
|
|
CA1902694 rs764594599 |
67 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1902692 rs575671552 |
71 | I>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs761580913 CA1902693 |
71 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA348872485 rs1407959561 |
78 | T>S | No |
ClinGen gnomAD |
|
|
CA1902666 rs748211534 |
82 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1272642255 CA348872406 |
88 | R>H | No |
ClinGen Ensembl |
|
|
CA348872399 rs1222381944 |
89 | P>H | No |
ClinGen Ensembl |
|
|
rs755197296 CA1902664 |
90 | L>V | No |
ClinGen ExAC gnomAD |
|
|
COSM175652 CA1902663 rs747141041 |
95 | S>L | Variant assessed as Somatic; 0.0 impact. large_intestine breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA348872354 rs758650254 |
96 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1231663472 CA348872349 |
97 | A>D | No |
ClinGen TOPMed |
|
|
rs1355458423 CA348872352 |
97 | A>T | No |
ClinGen TOPMed |
|
|
CA1902658 rs372548276 |
105 | S>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1347986827 CA348872280 |
107 | P>R | No |
ClinGen TOPMed |
|
|
rs753396212 CA1902657 |
107 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs752248279 CA1902654 |
112 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs367957253 CA1902653 |
115 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1178184010 CA348872218 |
116 | K>N | No |
ClinGen TOPMed |
|
|
CA348872193 rs1448290511 |
117 | W>C | No |
ClinGen gnomAD |
|
|
CA58434716 rs1017439234 |
119 | G>C | No |
ClinGen Ensembl |
|
|
CA58434715 rs959552606 |
119 | G>V | No |
ClinGen TOPMed |
|
|
CA348872180 rs1411768721 |
120 | E>K | No |
ClinGen TOPMed |
|
|
CA58434714 rs1007361820 |
121 | I>T | No |
ClinGen Ensembl |
|
|
CA58434713 rs1033868173 |
122 | Q>P | No |
ClinGen TOPMed |
|
|
rs777453229 CA1902634 |
125 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA1902633 rs755543766 |
126 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348872113 rs1272286193 |
129 | K>R | No |
ClinGen gnomAD |
|
|
rs142625334 CA58434712 |
131 | L>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP NCI-TCGA TOPMed gnomAD |
| TCGA novel | 131 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA348872082 rs1214214463 |
134 | G>S | No |
ClinGen gnomAD |
|
|
rs751315149 CA1902629 |
137 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1226352833 CA348872042 |
140 | R>G | No |
ClinGen gnomAD |
|
|
COSM716445 CA1902628 rs766103665 |
140 | R>Q | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs540244240 CA1902626 |
142 | D>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA348872015 rs1294665313 |
144 | S>N | No |
ClinGen gnomAD |
|
|
rs1048150963 CA58434710 |
145 | T>K | No |
ClinGen TOPMed gnomAD |
|
|
CA1902623 rs775841994 |
147 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA348871986 rs1390242501 |
149 | L>I | No |
ClinGen gnomAD |
|
|
CA1902621 rs746037801 |
151 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA348871971 rs746037801 |
151 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA348871965 rs1339441930 |
152 | H>Y | No |
ClinGen TOPMed |
|
|
CA348871958 rs1266537573 |
153 | R>G | No |
ClinGen gnomAD |
|
|
rs779437802 CA1902620 |
153 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA348871946 rs1489304179 |
154 | Q>H | No |
ClinGen gnomAD |
|
|
rs145448904 COSM145874 CA1902619 |
155 | T>M | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA1902617 rs778398680 |
156 | P>T | No |
ClinGen ExAC |
|
|
CA348871906 rs1223888524 |
161 | Q>E | No |
ClinGen TOPMed |
|
|
rs546539467 CA1902604 |
163 | A>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1902603 rs774679742 |
165 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749653637 CA1902601 |
166 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs771468851 CA1902602 |
166 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749653637 CA58434573 |
166 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA1902600 rs773921122 |
168 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs200146323 CA1902599 |
172 | A>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA348871822 rs1306694910 |
172 | A>P | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 172 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA348871803 rs1558868730 |
175 | I>V | No |
ClinGen Ensembl |
|
|
rs536121901 CA348871796 |
176 | E>* | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA58434571 COSM3836975 rs536121901 |
176 | E>K | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
|
CA1902595 rs746704792 |
177 | C>R | No |
ClinGen ExAC |
|
|
CA348871788 rs1400866920 |
177 | C>S | No |
ClinGen gnomAD |
|
|
rs1430293512 CA348871766 |
180 | L>F | No |
ClinGen gnomAD |
|
|
rs978242755 CA58434570 |
182 | S>L | No |
ClinGen TOPMed |
|
|
rs978242755 CA348871752 |
182 | S>W | No |
ClinGen TOPMed |
|
|
CA348871723 rs1192595445 |
186 | V>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA1902591 rs144867114 |
186 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1902590 rs757158401 |
187 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs753763478 CA1902589 |
191 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA1902587 rs765035010 |
192 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765035010 CA348871686 |
192 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763199337 CA1902584 |
196 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1331584919 CA348871649 |
198 | V>I | No |
ClinGen gnomAD |
|
|
CA348871636 rs1291894641 |
199 | N>K | No |
ClinGen gnomAD |
|
|
rs1242141817 CA348871635 |
200 | K>Q | No |
ClinGen gnomAD |
|
|
CA1902581 rs777165494 |
201 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777165494 CA1902580 |
201 | T>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777165494 CA348871624 |
201 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 202 | N>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768101389 CA1902579 |
202 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 202 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1902577 rs151069801 |
205 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs745467973 CA1902575 |
207 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA1902576 rs758054007 |
207 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1902574 rs778598169 |
210 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348871562 rs1369227927 |
211 | Q>E | No |
ClinGen gnomAD |
|
|
CA348871550 rs1189903110 |
212 | R>S | No |
ClinGen gnomAD |
|
|
rs532782714 CA58434569 |
213 | A>T | No |
ClinGen 1000Genomes TOPMed |
|
|
rs777749153 CA1902571 |
216 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348871505 rs1180754677 |
219 | H>Q | No |
ClinGen TOPMed |
|
|
CA348871502 rs1466682774 |
220 | M>V | No |
ClinGen gnomAD |
|
|
CA348871494 rs1364169705 |
221 | P>A | No |
ClinGen TOPMed |
|
|
CA1902570 rs755171874 |
222 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA348871489 rs1410266235 |
222 | S>R | No |
ClinGen gnomAD |
|
|
CA348871478 rs1573951640 |
223 | R>K | No |
ClinGen Ensembl |
|
|
CA1902567 rs763288592 |
224 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 225 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1362395691 CA348871466 |
225 | E>V | No |
ClinGen gnomAD |
|
|
CA1902566 rs750617672 |
226 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762217114 CA1902564 |
227 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1404494909 CA348871433 |
231 | T>A | No |
ClinGen gnomAD |
|
|
CA1902561 rs761105485 |
231 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771524793 CA1902559 |
232 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs1389618167 CA348871414 |
234 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
rs745554710 CA1902558 |
235 | K>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 236 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA348871386 rs1251632260 |
238 | A>T | No |
ClinGen gnomAD |
|
|
CA1902556 rs770685295 |
238 | A>V | No |
ClinGen ExAC gnomAD |
No associated diseases with P61587
1 regional properties for P61587
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 24 - 167 | IPR005225 |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cortical cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures in the cell cortex, i.e. just beneath the plasma membrane. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| small GTPase mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
37 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q00246 | RHO4 | GTP-binding protein RHO4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P61585 | RHOA | Transforming protein RhoA | Bos taurus (Bovine) | PR |
| Q1RMJ6 | RHOC | Rho-related GTP-binding protein RhoC | Bos taurus (Bovine) | PR |
| Q17QI8 | Rhov | Rho-related GTP-binding protein RhoV | Bos taurus (Bovine) | PR |
| Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB | Bos taurus (Bovine) | PR |
| Q2HJ68 | RND1 | Rho-related GTP-binding protein Rho6 | Bos taurus (Bovine) | PR |
| Q9PSX7 | RHOC | Rho-related GTP-binding protein RhoC | Gallus gallus (Chicken) | PR |
| P48148 | Rho1 | Ras-like GTP-binding protein Rho1 | Drosophila melanogaster (Fruit fly) | PR |
| O94955 | RHOBTB3 | Rho-related BTB domain-containing protein 3 | Homo sapiens (Human) | EV |
| O94844 | RHOBTB1 | Rho-related BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| P08134 | RHOC | Rho-related GTP-binding protein RhoC | Homo sapiens (Human) | PR |
| P61586 | RHOA | Transforming protein RhoA | Homo sapiens (Human) | PR |
| P62745 | RHOB | Rho-related GTP-binding protein RhoB | Homo sapiens (Human) | PR |
| Q92730 | RND1 | Rho-related GTP-binding protein Rho6 | Homo sapiens (Human) | PR |
| Q96L33 | RHOV | Rho-related GTP-binding protein RhoV | Homo sapiens (Human) | PR |
| Q62159 | Rhoc | Rho-related GTP-binding protein RhoC | Mus musculus (Mouse) | PR |
| Q9QUI0 | Rhoa | Transforming protein RhoA | Mus musculus (Mouse) | PR |
| Q9DAK3 | Rhobtb1 | Rho-related BTB domain-containing protein 1 | Mus musculus (Mouse) | PR |
| Q9CTN4 | Rhobtb3 | Rho-related BTB domain-containing protein 3 | Mus musculus (Mouse) | SS |
| Q8VDU1 | Rhov | Rho-related GTP-binding protein RhoV | Mus musculus (Mouse) | PR |
| P62746 | Rhob | Rho-related GTP-binding protein RhoB | Mus musculus (Mouse) | PR |
| Q8BLR7 | Rnd1 | Rho-related GTP-binding protein Rho6 | Mus musculus (Mouse) | PR |
| P61588 | Rnd3 | Rho-related GTP-binding protein RhoE | Mus musculus (Mouse) | PR |
| O77683 | RND3 | Rho-related GTP-binding protein RhoE | Sus scrofa (Pig) | PR |
| P62747 | Rhob | Rho-related GTP-binding protein RhoB | Rattus norvegicus (Rat) | PR |
| Q9Z1Y0 | Rhov | Rho-related GTP-binding protein RhoV | Rattus norvegicus (Rat) | PR |
| P61589 | Rhoa | Transforming protein RhoA | Rattus norvegicus (Rat) | PR |
| Q6SA80 | Rnd3 | Rho-related GTP-binding protein RhoE | Rattus norvegicus (Rat) | PR |
| Q9SSX0 | RAC1 | Rac-like GTP-binding protein 1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z808 | RAC3 | Rac-like GTP-binding protein 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q68Y52 | RAC2 | Rac-like GTP-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q67VP4 | RAC4 | Rac-like GTP-binding protein 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q22038 | rho-1 | Ras-like GTP-binding protein rhoA | Caenorhabditis elegans | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKERRASQKL | SSKSIMDPNQ | NVKCKIVVVG | DSQCGKTALL | HVFAKDCFPE | NYVPTVFENY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TASFEIDTQR | IELSLWDTSG | SPYYDNVRPL | SYPDSDAVLI | CFDISRPETL | DSVLKKWKGE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IQEFCPNTKM | LLVGCKSDLR | TDVSTLVELS | NHRQTPVSYD | QGANMAKQIG | AATYIECSAL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QSENSVRDIF | HVATLACVNK | TNKNVKRNKS | QRATKRISHM | PSRPELSAVA | TDLRKDKAKS |
| CTVM |