P61006
Gene name |
RAB8A (MEL, RAB8) |
Protein name |
Ras-related protein Rab-8A |
Names |
Oncogene c-mel |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4218 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
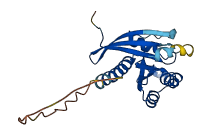
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

20 structures for P61006
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3QBT | X-ray | 200 A | A/C/E/G | 6-176 | PDB |
| 3TNF | X-ray | 250 A | A | 6-176 | PDB |
| 4LHV | X-ray | 195 A | A/B/C/D/E | 6-176 | PDB |
| 4LHW | X-ray | 155 A | A/B/C/D/E | 6-176 | PDB |
| 4LHX | X-ray | 305 A | A/B | 1-184 | PDB |
| 4LHY | X-ray | 310 A | A/B | 1-184 | PDB |
| 4LHZ | X-ray | 320 A | A/B | 1-184 | PDB |
| 4LI0 | X-ray | 330 A | A/B | 1-184 | PDB |
| 5SZI | X-ray | 285 A | A | 1-207 | PDB |
| 6RIR | X-ray | 177 A | A/B | 1-181 | PDB |
| 6SQ2 | X-ray | 168 A | A/B | 1-181 | PDB |
| 6STF | X-ray | 240 A | A/B/C/D/E | 5-176 | PDB |
| 6STG | X-ray | 250 A | A/B | 5-176 | PDB |
| 6WHE | X-ray | 173 A | A/B | 1-181 | PDB |
| 6YX5 | X-ray | 214 A | A | 6-176 | PDB |
| 6ZSI | X-ray | 191 A | A/B | 1-176 | PDB |
| 6ZSJ | X-ray | 200 A | A/B | 1-176 | PDB |
| 7BWT | X-ray | 230 A | B | 2-183 | PDB |
| 7LWB | X-ray | 190 A | A | 1-181 | PDB |
| AF-P61006-F1 | Predicted | AlphaFoldDB |
111 variants for P61006
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA404580493 rs1361108248 |
4 | T>S | No |
ClinGen gnomAD |
|
| TCGA novel | 5 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA404580498 rs1568317850 |
5 | Y>H | No |
ClinGen Ensembl |
|
|
CA9275585 rs768071919 |
14 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1181766596 CA404580577 |
17 | S>T | No |
ClinGen TOPMed |
|
|
CA404580594 rs1599393043 |
19 | V>G | No |
ClinGen Ensembl |
|
| TCGA novel | 26 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs751027770 CA9275591 |
31 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA404580686 rs1568317890 |
33 | F>Y | No |
ClinGen Ensembl |
|
|
rs781047043 CA9275593 |
34 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9275594 rs745787780 |
36 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA404580773 rs1208176747 |
44 | D>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1448892419 CA404580814 |
49 | T>I | No |
ClinGen gnomAD |
|
|
CA9275616 rs145971722 |
50 | I>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs139537285 CA9275617 |
50 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9275615 rs145971722 |
50 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1429377302 CA404580827 |
52 | L>F | No |
ClinGen TOPMed |
|
|
CA404580833 rs1599395321 |
53 | D>N | No |
ClinGen Ensembl |
|
|
CA404580843 rs1419063620 |
54 | G>S | No |
ClinGen TOPMed |
|
|
CA9275619 rs373314086 |
55 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs890623886 CA305901854 |
56 | R>I | No |
ClinGen Ensembl |
|
|
CA9275620 rs777976720 |
61 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404580895 rs1383515977 |
62 | W>R | No |
ClinGen gnomAD |
|
| TCGA novel | 63 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9275637 rs753016512 |
66 | G>C | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 66 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs758644794 CA9275638 |
67 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1213294850 CA404580950 |
68 | E>Q | No |
ClinGen gnomAD |
|
|
CA9275639 rs778028924 |
69 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1470442545 CA404580958 |
69 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1482944299 CA404580968 |
70 | F>L | No |
ClinGen gnomAD |
|
|
CA9275640 rs747042137 |
71 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs771353371 CA9275641 |
75 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs781750272 CA9275642 |
75 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 76 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 77 | Y>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA404581011 rs1432439795 |
78 | Y>H | No |
ClinGen gnomAD |
|
|
rs1473869285 CA404581065 |
84 | I>V | No |
ClinGen gnomAD |
|
|
CA404581073 rs1298136718 |
85 | M>L | No |
ClinGen gnomAD |
|
|
CA404581089 rs1308473301 |
87 | V>A | No |
ClinGen gnomAD |
|
|
CA305907834 rs1020965743 |
89 | D>N | No |
ClinGen Ensembl |
|
|
CA9275672 rs775954889 |
92 | N>T | No |
ClinGen ExAC gnomAD |
|
|
rs1049101274 CA305907836 |
93 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA9275674 rs765198343 |
97 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775438230 CA9275675 |
98 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA305907849 rs887364471 |
100 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs967148175 CA305907846 |
100 | R>W | No |
ClinGen Ensembl |
|
|
rs763790113 CA9275677 |
103 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA9275678 rs751842773 |
104 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA9275679 rs757394695 |
105 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1004305778 CA305907890 |
106 | I>T | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 107 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9275681 rs750406773 |
108 | E>Q | No |
ClinGen ExAC |
|
|
CA9275724 rs758754074 |
109 | H>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1421471379 CA404581259 |
110 | A>G | No |
ClinGen TOPMed |
|
|
CA9275726 COSM1680771 rs750145818 |
110 | A>T | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA404581270 rs1360547786 |
112 | A>E | No |
ClinGen TOPMed |
|
| TCGA novel | 112 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs949829005 CA305909467 |
114 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA305909472 rs1045591736 |
115 | E>K | No |
ClinGen TOPMed |
|
|
CA404581302 rs1424732985 |
117 | M>V | No |
ClinGen TOPMed |
|
|
CA404581311 rs1317165266 |
118 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1324362837 CA404581322 |
119 | L>R | No |
ClinGen gnomAD |
|
|
CA404581336 rs1333993671 |
121 | N>K | No |
ClinGen gnomAD |
|
|
rs1599398800 CA404581346 |
123 | C>R | No |
ClinGen Ensembl |
|
|
CA404581348 rs1275080931 |
123 | C>Y | No |
ClinGen gnomAD |
|
|
rs778785985 CA9275731 |
126 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9275732 rs748088384 |
128 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA305909509 rs369762374 CA9275733 |
128 | K>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA305909500 rs748088384 |
128 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1474491071 CA404581417 |
133 | K>R | No |
ClinGen gnomAD |
|
|
rs771073154 CA404581431 |
135 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9275736 rs771073154 |
135 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747264307 CA9275735 COSM1391240 |
135 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA9275737 CA404581452 rs776705625 |
138 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs768854359 CA9275778 |
142 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA305909832 rs765205828 |
142 | D>N | No |
ClinGen Ensembl |
|
|
rs775041655 COSM564969 CA9275779 |
143 | Y>C | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs372254577 CA305909850 |
146 | K>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA404581512 rs1267675900 |
146 | K>R | No |
ClinGen gnomAD |
|
|
rs772587714 CA9275781 |
148 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1164220285 CA404581554 |
152 | A>T | No |
ClinGen gnomAD |
|
|
CA305909877 rs145305113 |
155 | N>S | No |
ClinGen ESP TOPMed |
|
|
CA404581587 rs1249972414 |
156 | I>M | No |
ClinGen gnomAD |
|
|
rs764886982 CA9275784 |
157 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404581595 rs1233045102 |
158 | V>M | No |
ClinGen TOPMed |
|
|
rs1380876949 CA404581613 |
160 | N>S | No |
ClinGen gnomAD |
|
|
CA404581643 rs1423406940 |
163 | F>I | No |
ClinGen TOPMed |
|
|
CA9275813 rs747266932 |
163 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1160073390 CA404581655 |
164 | T>I | No |
ClinGen TOPMed |
|
|
rs758396964 CA9275816 |
166 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA9275815 rs758396964 |
166 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1434484343 CA404581678 |
168 | D>A | No |
ClinGen gnomAD |
|
|
CA9275817 rs747441158 |
168 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA305911226 rs201751163 |
172 | K>Q | No |
ClinGen Ensembl |
|
|
rs748965804 CA9275820 |
173 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1363132075 CA404581747 |
176 | K>I | No |
ClinGen TOPMed |
|
|
CA9275842 rs747736923 |
180 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9275844 rs772560627 |
181 | S>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA404581804 rs1183018562 |
181 | S>R | No |
ClinGen gnomAD |
|
|
CA305913188 rs927338669 |
188 | G>A | No |
ClinGen TOPMed |
|
| TCGA novel | 188 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs760666638 CA9275845 |
189 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA305913192 rs558096021 |
190 | K>R | No |
ClinGen 1000Genomes gnomAD |
|
|
rs770935465 CA9275846 |
191 | I>V | No |
ClinGen ExAC |
|
|
CA305913208 rs769827222 |
192 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA9275847 rs769827222 |
192 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs765092264 CA9275849 |
196 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA404581956 rs1321147098 |
200 | S>G | No |
ClinGen gnomAD |
|
|
rs1175467248 CA404581962 |
200 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA404582026 rs1599401002 |
208 | L>G | No |
ClinGen Ensembl |
No associated diseases with P61006
1 regional properties for P61006
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 7 - 162 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
24 GO annotations of cellular component
| Name | Definition |
|---|---|
| centriole | A cellular organelle, found close to the nucleus in many eukaryotic cells, consisting of a small cylinder with microtubular walls, 300-500 nm long and 150-250 nm in diameter. It contains nine short, parallel, peripheral microtubular fibrils, each fibril consisting of one complete microtubule fused to two incomplete microtubules. Cells usually have two centrioles, lying at right angles to each other. At division, each pair of centrioles generates another pair and the twin pairs form the pole of the mitotic spindle. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| ciliary basal body | A membrane-tethered, short cylindrical array of microtubules and associated proteins found at the base of a eukaryotic cilium (also called flagellum) that is similar in structure to a centriole and derives from it. The cilium basal body is the site of assembly and remodelling of the cilium and serves as a nucleation site for axoneme growth. As well as anchoring the cilium, it is thought to provide a selective gateway regulating the entry of ciliary proteins and vesicles by intraflagellar transport. |
| ciliary base | Area of the cilium (also called flagellum) where the basal body and the axoneme are anchored to the plasma membrane. The ciliary base encompasses the distal part of the basal body, transition fibers and transition zone and is structurally and functionally very distinct from the rest of the cilium. In this area proteins are sorted and filtered before entering the cilium, and many ciliary proteins localize specifically to this area. |
| ciliary membrane | The portion of the plasma membrane surrounding a cilium. |
| cilium | A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface and of some cytoplasmic parts. Each cilium is largely bounded by an extrusion of the cytoplasmic (plasma) membrane, and contains a regular longitudinal array of microtubules, anchored to a basal body. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| non-motile cilium | A cilium which may have a variable array of axonemal microtubules but does not contain molecular motors. |
| phagocytic vesicle | A membrane-bounded intracellular vesicle that arises from the ingestion of particulate material by phagocytosis. |
| phagocytic vesicle membrane | The lipid bilayer surrounding a phagocytic vesicle. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| recycling endosome membrane | The lipid bilayer surrounding a recycling endosome. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| trans-Golgi network membrane | The lipid bilayer surrounding any of the compartments that make up the trans-Golgi network. |
| trans-Golgi network transport vesicle | A vesicle that mediates transport between the trans-Golgi network and other parts of the cell. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| myosin V binding | Binding to a class V myosin; myosin V is a dimeric molecule involved in intracellular transport. |
| protein tyrosine kinase binding | Binding to protein tyrosine kinase. |
| small GTPase binding | Binding to a small monomeric GTPase. |
18 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| axonogenesis | De novo generation of a long process of a neuron, including the terminal branched region. Refers to the morphogenesis or creation of shape or form of the developing axon, which carries efferent (outgoing) action potentials from the cell body towards target cells. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cilium assembly | The assembly of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole. |
| endocytic recycling | The directed movement of membrane-bounded vesicles from endosomes back to the plasma membrane, a trafficking pathway that promotes the recycling of internalized transmembrane proteins. |
| Golgi organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the Golgi apparatus. |
| Golgi vesicle fusion to target membrane | The joining of the lipid bilayer membrane around a Golgi transport vesicle to the target lipid bilayer membrane. |
| neurotransmitter receptor transport to postsynaptic membrane | The directed movement of neurotransmitter receptor to the postsynaptic membrane in transport vesicles. |
| neurotransmitter receptor transport, endosome to postsynaptic membrane | The directed movement of neurotransmitter receptor from the postsynaptic endosome to the postsynaptic membrane in transport vesicles. |
| protein localization to cilium | A process in which a protein is transported to, or maintained in, a location within a cilium. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein secretion | The controlled release of proteins from a cell. |
| regulation of autophagy | Any process that modulates the frequency, rate or extent of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| regulation of long-term neuronal synaptic plasticity | A process that modulates long-term neuronal synaptic plasticity, the ability of neuronal synapses to change long-term as circumstances require. Long-term neuronal synaptic plasticity generally involves increase or decrease in actual synapse numbers. |
| regulation of protein transport | Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| vesicle docking involved in exocytosis | The initial attachment of a vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the vesicle and the target membrane, that contributes to exocytosis. |
| vesicle-mediated transport in synapse | Any vesicle-mediated transport that occurs in a synapse. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P07560 | SEC4 | Ras-related protein SEC4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q2HJI8 | RAB8B | Ras-related protein Rab-8B | Bos taurus (Bovine) | PR |
| Q1RMR4 | RAB15 | Ras-related protein Rab-15 | Bos taurus (Bovine) | PR |
| A4FV54 | RAB8A | Ras-related protein Rab-8A | Bos taurus (Bovine) | PR |
| Q5F470 | RAB8A | Ras-related protein Rab-8A | Gallus gallus (Chicken) | PR |
| P61007 | RAB8A | Ras-related protein Rab-8A | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P0C0E4 | RAB40AL | Ras-related protein Rab-40A-like | Homo sapiens (Human) | PR |
| Q8WXH6 | RAB40A | Ras-related protein Rab-40A | Homo sapiens (Human) | PR |
| Q12829 | RAB40B | Ras-related protein Rab-40B | Homo sapiens (Human) | PR |
| Q96S21 | RAB40C | Ras-related protein Rab-40C | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q92930 | RAB8B | Ras-related protein Rab-8B | Homo sapiens (Human) | PR |
| P59190 | RAB15 | Ras-related protein Rab-15 | Homo sapiens (Human) | PR |
| Q8K386 | Rab15 | Ras-related protein Rab-15 | Mus musculus (Mouse) | PR |
| P61028 | Rab8b | Ras-related protein Rab-8B | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q9DD03 | Rab13 | Ras-related protein Rab-13 | Mus musculus (Mouse) | PR |
| Q8CB87 | Rab44 | Ras-related protein Rab-44 | Mus musculus (Mouse) | PR |
| Q8VHP8 | Rab40b | Ras-related protein Rab-40B | Mus musculus (Mouse) | PR |
| Q8VHQ4 | Rab40c | Ras-related protein Rab-40C | Mus musculus (Mouse) | PR |
| P55258 | Rab8a | Ras-related protein Rab-8A | Mus musculus (Mouse) | PR |
| P35289 | Rab15 | Ras-related protein Rab-15 | Rattus norvegicus (Rat) | PR |
| P70550 | Rab8b | Ras-related protein Rab-8B | Rattus norvegicus (Rat) | PR |
| P35284 | Rab12 | Ras-related protein Rab-12 | Rattus norvegicus (Rat) | PR |
| P35281 | Rab10 | Ras-related protein Rab-10 | Rattus norvegicus (Rat) | PR |
| P35280 | Rab8a | Ras-related protein Rab-8A | Rattus norvegicus (Rat) | PR |
| O24466 | RABE1A | Ras-related protein RABE1a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SF91 | RABE1E | Ras-related protein RABE1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LZD4 | RABE1D | Ras-related protein RABE1d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAKTYDYLFK | LLLIGDSGVG | KTCVLFRFSE | DAFNSTFIST | IGIDFKIRTI | ELDGKRIKLQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IWDTAGQERF | RTITTAYYRG | AMGIMLVYDI | TNEKSFDNIR | NWIRNIEEHA | SADVEKMILG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NKCDVNDKRQ | VSKERGEKLA | LDYGIKFMET | SAKANINVEN | AFFTLARDIK | AKMDKKLEGN |
| 190 | 200 | ||||
| SPQGSNQGVK | ITPDQQKRSS | FFRCVLL |