P60842
Gene name |
EIF4A1 (DDX2A, EIF4A) |
Protein name |
Eukaryotic initiation factor 4A-I |
Names |
eIF-4A-I, eIF4A-I, ATP-dependent RNA helicase eIF4A-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1973 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
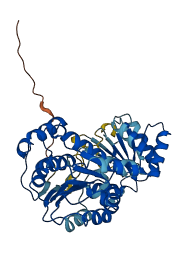
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

11 structures for P60842
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2G9N | X-ray | 225 A | A/B | 20-238 | PDB |
| 2ZU6 | X-ray | 280 A | A/C/D/F | 20-406 | PDB |
| 3EIQ | X-ray | 350 A | A/D | 1-406 | PDB |
| 5ZBZ | X-ray | 131 A | A | 20-238 | PDB |
| 5ZC9 | X-ray | 200 A | A | 19-406 | PDB |
| 6ZMW | EM | 370 A | j | 1-406 | PDB |
| 7PPZ | X-ray | 252 A | B | 20-406 | PDB |
| 7PQ0 | X-ray | 300 A | B | 20-406 | PDB |
| 8HUJ | EM | 376 A | A | 2-406 | PDB |
| 8OZ0 | EM | 350 A | 1/3 | 1-406 | PDB |
| AF-P60842-F1 | Predicted | AlphaFoldDB |
126 variants for P60842
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA397803592 rs139517056 |
2 | S>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs139517056 CA8351687 |
2 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8351688 rs779627306 |
3 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA287456596 rs891239542 |
3 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8351690 rs768379368 |
4 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200951325 CA8351692 |
4 | S>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs759758160 CA8351695 |
5 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA397803628 rs149681191 |
5 | Q>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8351694 rs149681191 |
5 | Q>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs149681191 CA8351693 |
5 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs950953435 CA287456646 |
6 | D>G | No |
ClinGen TOPMed |
|
|
CA8351696 rs199974690 COSM707102 |
7 | S>F | lung Variant assessed as Somatic; 0.0 impact. large_intestine urinary_tract [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA397803652 rs1272295986 |
7 | S>P | No |
ClinGen gnomAD |
|
|
rs199974690 CA287456653 |
7 | S>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
rs928267905 COSM244232 CA287456665 |
8 | R>* | endometrium prostate [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs560471589 CA8351698 |
8 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs372189155 CA8351754 |
18 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 20 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1258002273 CA397804797 |
24 | E>K | No |
ClinGen TOPMed |
|
|
CA397804914 rs1233262122 |
25 | S>C | No |
ClinGen gnomAD |
|
|
CA8351797 rs749545098 |
25 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA397804921 rs1277319699 |
26 | N>D | No |
ClinGen TOPMed |
|
|
CA397804959 rs1364069393 |
28 | N>D | No |
ClinGen TOPMed |
|
| TCGA novel | 28 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA397804993 rs1233986181 |
29 | E>D | No |
ClinGen gnomAD |
|
|
rs11552651 CA287458835 |
30 | I>F | No |
ClinGen Ensembl |
|
|
rs779337219 CA8351799 |
33 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA397805051 rs1415163964 |
33 | S>R | No |
ClinGen gnomAD |
|
|
rs1485050805 CA397805064 |
34 | F>S | No |
ClinGen gnomAD |
|
| TCGA novel | 35 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1270678661 CA397805095 |
36 | D>A | No |
ClinGen TOPMed |
|
|
rs1419941523 CA397805163 |
40 | S>A | No |
ClinGen gnomAD |
|
|
rs1036532248 CA287458865 |
40 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| TCGA novel | 41 | E>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA397805197 rs1455059798 |
42 | S>F | No |
ClinGen gnomAD |
|
|
CA397805228 rs1400990316 |
45 | R>C | No |
ClinGen gnomAD |
|
|
CA397805230 rs1414585747 |
45 | R>H | No |
ClinGen gnomAD |
|
| TCGA novel | 45 | R>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs762799240 CA8351809 |
55 | P>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 55 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs766144308 CA8351810 |
57 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs11552638 CA287458928 |
61 | R>L | No |
ClinGen Ensembl |
|
|
CA397805448 rs1597847982 |
62 | A>T | No |
ClinGen Ensembl |
|
| TCGA novel | 63 | I>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8351813 rs764244678 |
66 | C>S | No |
ClinGen ExAC gnomAD |
|
|
rs565470380 CA8351815 |
68 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA397805803 rs1250791633 |
70 | Y>* | No |
ClinGen gnomAD |
|
|
rs11552634 CA287459562 |
75 | Q>* | No |
ClinGen Ensembl |
|
|
CA397806088 rs1280149606 |
77 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA287459586 rs11552641 |
84 | A>S | No |
ClinGen Ensembl |
|
|
CA287459595 rs11552636 |
89 | S>A | No |
ClinGen Ensembl |
|
|
CA8351891 rs768559825 |
90 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA8351890 rs768559825 |
90 | I>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 94 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs374811712 CA397806580 |
95 | E>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374811712 CA8351893 |
95 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1234645199 CA397806635 |
97 | D>G | No |
ClinGen gnomAD |
|
|
rs145214315 CA8351894 |
100 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8351896 rs774233496 |
107 | A>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 110 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 111 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1466821541 CA397809261 |
121 | M>I | No |
ClinGen gnomAD |
|
| TCGA novel | 122 | A>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA397809327 rs1429277539 |
124 | G>R | No |
ClinGen TOPMed |
|
|
CA287462108 rs1002645516 |
126 | Y>S | No |
ClinGen Ensembl |
|
|
rs1567734571 CA397809425 COSM984774 |
127 | M>T | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA8351972 rs767102356 |
127 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA397809448 rs1170380404 |
128 | G>S | No |
ClinGen gnomAD |
|
|
rs760193070 CA8351974 |
129 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs760193070 CA397809467 |
129 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 132 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs148339522 CA8351977 |
133 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA287462153 rs985461314 |
138 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs1229174572 CA397809826 |
139 | N>S | No |
ClinGen TOPMed |
|
|
CA397809846 rs1319861944 |
140 | V>M | No |
ClinGen gnomAD |
|
|
CA8351983 rs756236929 |
141 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1489015522 CA397810119 |
148 | Q>H | No |
ClinGen gnomAD |
|
|
CA287462156 rs11552632 |
148 | Q>L | No |
ClinGen Ensembl |
|
|
rs1186255431 CA397810135 |
149 | M>V | No |
ClinGen gnomAD |
|
|
rs1171527049 CA397810341 |
155 | I>V | No |
ClinGen gnomAD |
|
|
CA397810758 rs1350615995 |
168 | R>Q | No |
ClinGen gnomAD |
|
|
rs1465951505 CA397811349 |
174 | K>R | No |
ClinGen gnomAD |
|
|
rs1301237658 CA397811368 |
175 | Y>N | No |
ClinGen gnomAD |
|
|
CA397811435 rs1330933425 |
177 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 177 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1374895713 CA397811502 |
178 | M>I | No |
ClinGen gnomAD |
|
| TCGA novel | 178 | M>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 185 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA397812034 rs1177832426 |
192 | F>C | No |
ClinGen gnomAD |
|
|
CA397812087 rs1246790916 |
194 | D>N | No |
ClinGen TOPMed |
|
| TCGA novel | 204 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 219 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs865904208 | 233 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1418798284 | 247 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1283063684 | 251 | I>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 253 | V>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 268 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 284 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA397815719 rs1480750944 |
291 | K>M | No |
ClinGen TOPMed |
|
| TCGA novel | 292 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1317590696 CA397815733 |
292 | M>L | No |
ClinGen gnomAD |
|
|
rs1567735382 CA397815761 |
293 | H>R | No |
ClinGen Ensembl |
|
|
CA8352178 rs758502373 |
295 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA397815850 rs1438216218 |
299 | V>I | No |
ClinGen gnomAD |
|
|
CA397815891 rs1244960491 |
301 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA287462939 rs112659619 |
302 | M>I | No |
ClinGen Ensembl |
|
|
CA8352201 rs752935351 |
306 | M>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 311 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs186439250 CA287463038 |
313 | V>M | No |
ClinGen 1000Genomes gnomAD |
|
| TCGA novel | 323 | S>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 330 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8352209 rs775034603 |
331 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs61730294 CA287463284 |
344 | V>L | No |
ClinGen Ensembl |
|
|
CA397817783 rs1156365059 |
347 | Y>C | No |
ClinGen gnomAD |
|
|
CA397817763 rs1296084728 |
347 | Y>H | No |
ClinGen gnomAD |
|
| TCGA novel | 365 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8352339 rs761383765 |
373 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1363920037 CA397818981 |
375 | M>V | No |
ClinGen gnomAD |
|
|
rs1274842003 CA397819122 |
379 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs368951361 CA8352342 |
380 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1567735983 CA397819151 |
381 | K>T | No |
ClinGen Ensembl |
|
|
rs754812366 CA8352345 |
384 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs781174047 CA8352346 |
385 | R>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 390 | F>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746054327 CA8352350 |
395 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1406896419 CA397819668 |
401 | N>S | No |
ClinGen gnomAD |
No associated diseases with P60842
6 regional properties for P60842
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | ATP-dependent RNA helicase DEAD-box, conserved site | 180 - 188 | IPR000629 |
| domain | Helicase, C-terminal | 245 - 406 | IPR001650 |
| domain | DEAD/DEAH box helicase domain | 57 - 221 | IPR011545 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 51 - 249 | IPR014001 |
| domain | RNA helicase, DEAD-box type, Q motif | 32 - 60 | IPR014014 |
| domain | ATP-dependent RNA helicase eIF4A, DEAD-box helicase domain | 34 - 235 | IPR044728 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| eukaryotic translation initiation factor 4F complex | The eukaryotic translation initiation factor 4F complex is composed of eIF4E, eIF4A and eIF4G; it is involved in the recognition of the mRNA cap, ATP-dependent unwinding of the 5'-terminal secondary structure and recruitment of the mRNA to the ribosome. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
10 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| double-stranded RNA binding | Binding to double-stranded RNA. |
| helicase activity | Catalysis of the reaction: ATP + H2O = ADP + phosphate, to drive the unwinding of a DNA or RNA helix. |
| mRNA binding | Binding to messenger RNA (mRNA), an intermediate molecule between DNA and protein. mRNA includes UTR and coding sequences, but does not contain introns. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA cap binding | Binding to a 7-methylguanosine (m7G) group or derivative located at the 5' end of an RNA molecule. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
| translation factor activity, RNA binding | Functions during translation by binding to RNA during polypeptide synthesis at the ribosome. |
| translation initiation factor activity | Functions in the initiation of ribosome-mediated translation of mRNA into a polypeptide. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cytoplasmic translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein in the cytoplasm. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
| translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P41378 | Eukaryotic initiation factor 4A | Triticum aestivum (Wheat) | PR | |
| P10081 | TIF2 | ATP-dependent RNA helicase eIF4A | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q3SZ65 | EIF4A2 | Eukaryotic initiation factor 4A-II | Bos taurus (Bovine) | PR |
| Q3SZ54 | EIF4A1 | Eukaryotic initiation factor 4A-I | Bos taurus (Bovine) | PR |
| Q8JFP1 | EIF4A2 | Eukaryotic initiation factor 4A-II | Gallus gallus (Chicken) | PR |
| A5A6N4 | EIF4A1 | Eukaryotic initiation factor 4A-I | Pan troglodytes (Chimpanzee) | PR |
| Q8NHQ9 | DDX55 | ATP-dependent RNA helicase DDX55 | Homo sapiens (Human) | PR |
| Q14240 | EIF4A2 | Eukaryotic initiation factor 4A-II | Homo sapiens (Human) | PR |
| Q9Y6V7 | DDX49 | Probable ATP-dependent RNA helicase DDX49 | Homo sapiens (Human) | PR |
| Q9H0S4 | DDX47 | Probable ATP-dependent RNA helicase DDX47 | Homo sapiens (Human) | PR |
| Q8TDD1 | DDX54 | ATP-dependent RNA helicase DDX54 | Homo sapiens (Human) | PR |
| Q9GZR7 | DDX24 | ATP-dependent RNA helicase DDX24 | Homo sapiens (Human) | PR |
| Q92499 | DDX1 | ATP-dependent RNA helicase DDX1 | Homo sapiens (Human) | PR |
| Q9NY93 | DDX56 | Probable ATP-dependent RNA helicase DDX56 | Homo sapiens (Human) | PR |
| Q9NUL7 | DDX28 | Probable ATP-dependent RNA helicase DDX28 | Homo sapiens (Human) | PR |
| Q7L014 | DDX46 | Probable ATP-dependent RNA helicase DDX46 | Homo sapiens (Human) | PR |
| Q41741 | Eukaryotic initiation factor 4A | Zea mays (Maize) | PR | |
| P10630 | Eif4a2 | Eukaryotic initiation factor 4A-II | Mus musculus (Mouse) | PR |
| P60843 | Eif4a1 | Eukaryotic initiation factor 4A-I | Mus musculus (Mouse) | PR |
| Q5RKI1 | Eif4a2 | Eukaryotic initiation factor 4A-II | Rattus norvegicus (Rat) | PR |
| Q6Z2Z4 | Os02g0146600 | Eukaryotic initiation factor 4A-3 | Oryza sativa subsp japonica (Rice) | PR |
| P35683 | Os06g0701100 | Eukaryotic initiation factor 4A-1 | Oryza sativa subsp japonica (Rice) | PR |
| P27639 | inf-1 | Eukaryotic initiation factor 4A | Caenorhabditis elegans | PR |
| Q3E9C3 | RH58 | DEAD-box ATP-dependent RNA helicase 58, chloroplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9CAI7 | TIF4A-3 | Eukaryotic initiation factor 4A-3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41376 | EIF4A1 | Eukaryotic initiation factor 4A-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41377 | TIF4A-2 | Eukaryotic initiation factor 4A-2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSASQDSRSR | DNGPDGMEPE | GVIESNWNEI | VDSFDDMNLS | ESLLRGIYAY | GFEKPSAIQQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RAILPCIKGY | DVIAQAQSGT | GKTATFAISI | LQQIELDLKA | TQALVLAPTR | ELAQQIQKVV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MALGDYMGAS | CHACIGGTNV | RAEVQKLQME | APHIIVGTPG | RVFDMLNRRY | LSPKYIKMFV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LDEADEMLSR | GFKDQIYDIF | QKLNSNTQVV | LLSATMPSDV | LEVTKKFMRD | PIRILVKKEE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTLEGIRQFY | INVEREEWKL | DTLCDLYETL | TITQAVIFIN | TRRKVDWLTE | KMHARDFTVS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AMHGDMDQKE | RDVIMREFRS | GSSRVLITTD | LLARGIDVQQ | VSLVINYDLP | TNRENYIHRI |
| 370 | 380 | 390 | 400 | ||
| GRGGRFGRKG | VAINMVTEED | KRTLRDIETF | YNTSIEEMPL | NVADLI |