P55040
Gene name |
GEM (KIR) |
Protein name |
GTP-binding protein GEM |
Names |
GTP-binding mitogen-induced T-cell protein, RAS-like protein KIR |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2669 |
EC number |
|
Protein Class |
|
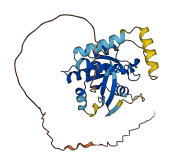
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for P55040
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CJW | X-ray | 210 A | A/B | 74-261 | PDB |
| 2G3Y | X-ray | 240 A | A | 62-249 | PDB |
| 2HT6 | X-ray | 240 A | A/B | 71-243 | PDB |
| AF-P55040-F1 | Predicted | AlphaFoldDB |
282 variants for P55040
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 1 | M>? | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746033873 CA4809949 |
2 | T>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 3 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4809947 rs779094271 |
6 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA4809946 rs771211990 |
7 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371700441 rs771211990 |
7 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371700446 rs1586066866 |
7 | T>P | No |
ClinGen Ensembl |
|
|
rs374909426 CA4809945 |
8 | M>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA371700437 rs1217047065 |
8 | M>V | No |
ClinGen Ensembl |
|
|
COSM1102553 rs777960854 CA4809944 |
9 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA4809943 rs756531627 |
9 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs556308745 CA4809942 |
11 | G>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809941 rs556308745 |
11 | G>D | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809939 rs370415265 |
14 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs542542829 CA4809940 |
14 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA371700374 rs370415265 |
14 | G>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA371700365 rs1167766631 |
15 | M>I | No |
ClinGen TOPMed |
|
|
rs34218332 CA181383042 |
15 | M>T | No |
ClinGen Ensembl |
|
|
CA371700372 rs1435182389 |
15 | M>V | No |
ClinGen TOPMed |
|
|
rs764949315 CA4809938 |
16 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761603767 CA4809937 |
17 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1171125278 CA371700347 |
17 | P>T | No |
ClinGen gnomAD |
|
|
CA4809936 rs753235715 |
20 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371700310 rs1189933908 |
20 | Q>R | No |
ClinGen gnomAD |
|
|
rs140549853 CA4809935 |
21 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
rs760461262 CA4809934 |
22 | W>C | No |
ClinGen ExAC gnomAD |
|
|
rs775511173 CA4809933 |
23 | S>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 28 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA371700198 rs1586066742 |
30 | H>Q | No |
ClinGen Ensembl |
|
|
rs774173558 CA4809930 |
30 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA181383009 rs1027914797 |
32 | M>I | No |
ClinGen Ensembl |
|
|
rs1586066729 CA371700163 |
33 | V>G | No |
ClinGen Ensembl |
|
|
rs1317636210 CA371700158 |
34 | Q>* | No |
ClinGen gnomAD |
|
|
rs771122242 CA4809929 |
34 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1563608593 CA371700147 |
35 | K>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA181383003 rs11539260 |
35 | K>I | No |
ClinGen Ensembl |
|
|
rs769925332 CA181382977 CA4809926 |
36 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371700136 rs1350688149 |
36 | E>K | No |
ClinGen TOPMed |
|
|
rs1378667182 CA371700119 |
38 | H>P | No |
ClinGen gnomAD |
|
|
rs1378667182 CA371700121 |
38 | H>R | No |
ClinGen gnomAD |
|
|
CA4809925 rs35262320 RCV000911488 |
38 | H>Y | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA4809924 rs781762421 |
39 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA4809923 rs755579538 |
40 | Y>D | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 41 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA371700096 rs1397815569 |
41 | S>R | No |
ClinGen gnomAD |
|
|
CA371700089 rs1586066645 |
42 | H>P | No |
ClinGen Ensembl |
|
|
CA181382962 rs35065846 |
42 | H>Q | No |
ClinGen TOPMed |
|
|
CA4809922 rs2170363 |
43 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs2170363 CA4809921 VAR_020097 |
43 | R>G | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs967988799 CA181382927 |
43 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs967988799 CA371700086 |
43 | R>P | No |
ClinGen TOPMed gnomAD |
|
|
rs2170363 CA371700087 |
43 | R>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA181382904 rs1017925631 |
44 | N>D | No |
ClinGen gnomAD |
|
|
rs1017925631 CA371700084 |
44 | N>H | No |
ClinGen gnomAD |
|
|
CA4809918 rs763453975 |
44 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs763453975 CA4809919 |
44 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA4809916 rs201351710 |
45 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809915 rs17847126 |
45 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs201351710 CA4809917 |
45 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1249472878 CA371700065 |
47 | S>F | No |
ClinGen gnomAD |
|
|
rs1184282847 CA371700062 |
48 | A>T | No |
ClinGen gnomAD |
|
|
CA4809913 rs774085906 |
49 | T>A | Variant assessed as Somatic; 4.621e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA4809912 rs766195976 |
49 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA371700056 rs774085906 |
49 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs147133582 CA4809910 |
50 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1586066522 CA371700036 |
52 | D>A | No |
ClinGen Ensembl |
|
|
rs935211262 CA181382881 |
53 | H>N | No |
ClinGen gnomAD |
|
|
CA371700029 rs1586066516 |
53 | H>P | No |
ClinGen Ensembl |
|
|
rs769234376 CA4809906 |
55 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA371700014 rs1295067536 |
55 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs956176521 CA181382840 |
56 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
COSM296578 CA4809904 rs780498125 |
56 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA371700005 rs1422864436 |
57 | S>N | No |
ClinGen gnomAD |
|
|
rs758803093 COSM1102552 CA4809902 |
59 | S>F | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA4809901 rs746276333 |
61 | D>A | No |
ClinGen ExAC gnomAD |
|
|
rs145616833 RCV000884716 CA4809900 |
61 | D>E | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA4809899 rs755764047 |
62 | S>F | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 66 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs767038807 CA4809897 |
66 | V>D | No |
ClinGen ExAC gnomAD |
|
|
CA371699940 rs1286096333 |
67 | I>T | No |
ClinGen gnomAD |
|
|
rs754873278 CA4809896 |
69 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809894 rs751506282 |
72 | G>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs762800000 CA4809892 |
74 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772917669 CA4809891 |
75 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371699891 rs772917669 |
75 | Y>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809890 rs765472126 |
76 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs550393204 CA181382732 |
77 | R>* | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA371699880 rs550393204 |
77 | R>G | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs761960990 CA4809889 |
77 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA371699876 rs1343671433 |
78 | V>M | No |
ClinGen gnomAD |
|
| TCGA novel | 79 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768860314 CA4809887 |
79 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1246178493 CA371699852 |
80 | L>P | No |
ClinGen TOPMed |
|
|
CA371699837 rs1197965766 |
81 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA4809885 rs776127220 |
82 | G>E | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 83 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA371699794 rs1186893760 |
84 | Q>H | No |
ClinGen TOPMed |
|
|
CA4809883 rs201303048 |
84 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs567967510 CA4809881 |
85 | G>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA4809882 rs139494082 |
85 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs139494082 CA371699787 |
85 | G>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA181382679 rs916486616 |
86 | V>L | No |
ClinGen Ensembl |
|
|
rs916486616 CA181382681 |
86 | V>M | No |
ClinGen Ensembl |
|
|
CA181382676 rs567771198 |
88 | K>R | No |
ClinGen Ensembl |
|
|
CA4809880 rs747832501 |
90 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs887416077 CA181382665 |
93 | N>D | No |
ClinGen TOPMed |
|
|
rs1341064482 CA371699688 |
93 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1047451181 CA181382654 |
98 | V>A | No |
ClinGen TOPMed |
|
|
rs754500839 CA371699606 |
99 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs754500839 CA4809878 |
99 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA371699611 rs1277426619 |
99 | H>Y | No |
ClinGen gnomAD |
|
|
rs1446048750 CA371699585 |
100 | D>E | No |
ClinGen gnomAD |
|
|
CA4809877 rs751132195 |
102 | M>R | No |
ClinGen ExAC gnomAD |
|
|
CA4809875 rs780005500 |
104 | S>C | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 104 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4809873 rs548062745 |
105 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs548062745 CA371699519 |
105 | D>Y | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs764970254 CA4809872 |
106 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA4809871 rs762025958 |
106 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA371699481 rs201792479 |
107 | E>D | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809869 rs527705763 |
107 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
rs527705763 CA371699493 |
107 | E>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA371699475 rs1182152545 |
108 | V>L | No |
ClinGen gnomAD |
|
|
CA181377633 rs201677240 |
113 | T>A | No |
ClinGen 1000Genomes |
|
|
CA4809854 rs757104245 |
113 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs762667312 CA4809853 |
116 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA371698548 rs762667312 |
116 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs1237330383 CA371698547 |
116 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA4809852 rs764380913 |
119 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA181377608 rs1042024547 |
119 | M>T | No |
ClinGen Ensembl |
|
|
rs530616234 CA181377592 |
120 | V>A | No |
ClinGen Ensembl |
|
|
rs761027449 CA4809851 |
120 | V>L | No |
ClinGen ExAC |
|
|
CA4809849 rs767526831 |
122 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs767526831 CA4809850 |
122 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs774736541 CA4809847 |
125 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1321156262 CA371698488 |
126 | T>A | No |
ClinGen gnomAD |
|
|
CA4809846 rs184919570 |
126 | T>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs773612061 CA371698459 |
131 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773612061 CA4809844 |
131 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1331101096 CA371698437 |
133 | W>* | No |
ClinGen gnomAD |
|
|
CA4809830 rs200005791 |
137 | G>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809829 rs751732240 |
138 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA4809827 rs763469017 |
139 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA371697712 rs763469017 |
139 | N>H | No |
ClinGen ExAC gnomAD |
|
|
rs1479417746 CA371697699 |
139 | N>K | No |
ClinGen gnomAD |
|
| TCGA novel | 139 | N>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1265172470 CA371697595 |
143 | H>Y | No |
ClinGen gnomAD |
|
|
rs1195962183 CA371697461 |
146 | C>* | No |
ClinGen gnomAD |
|
|
rs557922897 CA181377013 |
147 | M>T | No |
ClinGen gnomAD |
|
|
CA371697446 rs1488634712 |
147 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA4809826 rs773522304 |
148 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA4809823 rs774991627 |
150 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA371697353 rs1162622695 |
151 | D>G | No |
ClinGen TOPMed |
|
|
rs372249284 CA4809821 |
152 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1400113284 CA371697299 |
153 | Y>C | No |
ClinGen gnomAD |
|
|
rs1280600378 CA371697306 |
153 | Y>N | No |
ClinGen gnomAD |
|
| TCGA novel | 154 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA371697230 rs773969361 |
156 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs773969361 CA4809820 |
156 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs770754951 CA181376971 |
157 | Y>C | No |
ClinGen Ensembl |
|
|
CA4809819 rs770933590 CA371697164 |
158 | S>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1386177895 CA371697136 |
159 | I>T | No |
ClinGen TOPMed |
|
|
CA371697150 rs1563605554 |
159 | I>V | No |
ClinGen Ensembl |
|
|
rs1386530644 CA371697110 |
160 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs777669807 CA371697086 |
161 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA181376931 rs976230477 |
162 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
rs149283301 CA4809816 |
162 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs748020922 CA371697039 |
163 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748020922 CA4809815 COSM1102549 |
163 | A>V | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1425728424 CA371697020 |
164 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
CA371697015 rs1425728424 |
164 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs781236754 CA4809814 |
166 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751539968 CA4809813 |
168 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751539968 CA4809812 |
168 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809811 rs766455469 |
169 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA181376887 COSM198776 rs1011758304 |
172 | R>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs750953687 CA4809809 COSM1674013 |
172 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA371696853 rs1203460397 |
173 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
CA4809808 rs201731029 |
174 | Q>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs201731029 CA371696840 |
174 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs762054664 CA4809807 |
175 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs777120814 CA4809806 |
176 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371696812 rs777120814 |
176 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809804 rs759240105 |
176 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809805 COSM123341 rs759240105 |
176 | R>L | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1458575369 CA371696802 |
177 | R>G | No |
ClinGen TOPMed |
|
|
rs868545032 CA181376873 |
178 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs770594268 CA4809802 |
178 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1301234891 CA371696755 COSM1458699 |
179 | R>Q | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs577912523 CA4809801 |
179 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs74360159 CA181376843 |
181 | T>K | No |
ClinGen Ensembl |
|
|
CA371696717 rs1357438625 |
182 | E>K | No |
ClinGen TOPMed |
|
|
rs1450233215 CA371696697 |
183 | D>N | No |
ClinGen gnomAD |
|
|
CA371696644 rs1171364667 |
187 | I>L | No |
ClinGen gnomAD |
|
|
rs375183063 CA4809798 |
190 | G>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA371696574 rs1166246602 |
192 | K>R | No |
ClinGen gnomAD |
|
|
rs1473792037 CA371696555 |
193 | S>I | No |
ClinGen gnomAD |
|
|
rs1183193564 CA371696503 |
197 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA4809797 COSM239964 rs187593880 |
197 | R>W | pancreas endometrium Variant assessed as Somatic; 4.62e-05 impact. prostate [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs558611127 CA4809796 |
198 | C>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4809795 rs138582164 |
199 | R>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4809794 rs780063278 |
199 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4809793 rs758453208 |
203 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371696407 rs1356399304 |
204 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs778977780 CA4809773 |
208 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA371696229 rs1223035125 |
209 | C>F | No |
ClinGen gnomAD |
|
|
CA371696192 rs1167767811 |
212 | V>A | No |
ClinGen gnomAD |
|
|
rs754399008 CA4809771 |
212 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1437093428 CA371696153 |
215 | C>R | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 217 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4809770 rs138012458 |
218 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4809768 rs753149152 |
219 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1102548 CA4809767 rs766122201 |
220 | T>N | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA |
|
CA371696013 rs1362722619 |
225 | Q>R | No |
ClinGen gnomAD |
|
|
rs1160061065 CA371696006 |
226 | H>D | No |
ClinGen gnomAD |
|
|
CA4809766 rs762747900 |
227 | N>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 227 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs764837790 CA4809764 |
228 | V>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 228 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4809765 rs370285889 |
228 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 230 | E>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA181376042 rs921666967 |
231 | L>P | No |
ClinGen Ensembl |
|
|
rs1196682989 CA371695883 |
232 | F>S | No |
ClinGen gnomAD |
|
| TCGA novel | 234 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4809761 rs141761903 |
235 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs138147292 CA4809760 |
237 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA4809758 rs368821999 COSM1700368 |
237 | R>Q | skin [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1344821959 CA371695724 |
239 | V>A | No |
ClinGen gnomAD |
|
|
CA4809757 rs746095122 |
239 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs190907564 CA4809756 |
240 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4809755 rs771063860 |
240 | R>H | Variant assessed as Somatic; 4.619e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA371695706 rs771063860 |
240 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371695679 rs1563604832 |
242 | R>Q | No |
ClinGen Ensembl |
|
|
rs749374110 COSM2791371 CA4809754 |
242 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs200768882 CA4809752 |
243 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs778310132 CA4809753 |
243 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs141202489 CA4809749 |
247 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 248 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1586058486 CA371695543 |
248 | K>R | No |
ClinGen Ensembl |
|
|
CA181375966 rs867506754 |
250 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
CA371695465 rs1261805228 COSM198775 |
251 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs200775391 CA4809747 |
251 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs375369112 CA4809745 |
252 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs138265181 CA4809746 |
252 | R>W | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4809744 rs764080201 |
253 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs760620813 CA4809743 |
254 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs148957520 CA371695334 |
258 | R>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4809741 rs148957520 |
258 | R>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1416532243 CA371695317 |
259 | K>E | No |
ClinGen TOPMed |
|
|
rs774608878 CA4809739 |
260 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA371695297 rs759705261 |
260 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs759705261 CA4809740 |
260 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs771095990 CA4809738 |
261 | S>G | No |
ClinGen ExAC |
|
|
rs749379436 CA4809737 |
261 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs770363121 CA4809735 |
262 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA4809736 rs773218409 |
262 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA4809733 rs1313265181 |
263 | P>L | No |
ClinGen TOPMed |
|
|
CA371695221 rs1313265181 |
263 | P>R | No |
ClinGen TOPMed |
|
|
CA371695175 rs1304770745 |
266 | A>T | No |
ClinGen gnomAD |
|
|
rs748700800 CA4809732 |
266 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs755375862 CA4809730 |
268 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs145789382 CA4809729 COSM1102546 |
268 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs753332084 CA4809726 |
272 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs149831559 CA371694994 |
274 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs149831559 CA4809724 |
274 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 277 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs767490302 CA4809722 |
278 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs752653844 CA4809723 |
278 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA4809721 rs759294461 |
279 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs557181794 CA181375825 |
280 | N>T | No |
ClinGen 1000Genomes |
|
|
rs940281240 CA181375813 |
281 | M>V | No |
ClinGen gnomAD |
|
|
CA4809720 rs372371105 |
282 | A>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA371694880 rs1250641425 |
282 | A>T | No |
ClinGen gnomAD |
|
|
CA371694842 rs1213567190 |
284 | K>R | No |
ClinGen TOPMed |
|
|
CA4809719 rs766608259 COSM339538 |
285 | L>F | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA371694798 rs1179266588 |
287 | S>Y | No |
ClinGen TOPMed |
|
|
rs770019011 CA4809716 |
289 | S>P | No |
ClinGen ExAC gnomAD |
|
|
rs777248979 CA4809714 |
291 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs762037099 CA4809715 |
291 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA4809713 rs769356943 |
292 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA4809712 rs747387622 |
294 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA4809711 rs537389614 |
296 | L>F | No |
ClinGen 1000Genomes ExAC gnomAD |
No associated diseases with P55040
5 regional properties for P55040
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 444 - 719 | IPR000719 |
| domain | EGF-like calcium-binding domain | 336 - 363 | IPR001881 |
| active_site | Serine/threonine-protein kinase, active site | 566 - 578 | IPR008271 |
| domain | Wall-associated receptor kinase | 197 - 305 | IPR013695 |
| conserved_site | EGF-like calcium-binding, conserved site | 336 - 362 | IPR018097 |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasmic side of plasma membrane | The leaflet the plasma membrane that faces the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| mitotic spindle | A spindle that forms as part of mitosis. Mitotic and meiotic spindles contain distinctive complements of proteins associated with microtubules. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| spindle midzone | The area in the center of the spindle where the spindle microtubules from opposite poles overlap. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium channel regulator activity | Modulates the activity of a calcium channel. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| chromosome organization | A process that is carried out at the cellular level that results in the assembly, arrangement of constituent parts, or disassembly of chromosomes, structures composed of a very long molecule of DNA and associated proteins that carries hereditary information. This term covers covalent modifications at the molecular level as well as spatial relationships among the major components of a chromosome. |
| immune response | Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat. |
| metaphase plate congression | The alignment of chromosomes at the metaphase plate (spindle equator), a plane halfway between the poles of the spindle. |
| mitotic cell cycle | Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent. |
| negative regulation of high voltage-gated calcium channel activity | Any process that stops, prevents or reduces the frequency, rate or extent of high voltage-gated calcium channel activity. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTLNNVTMRQ | GTVGMQPQQQ | RWSIPADGRH | LMVQKEPHQY | SHRNRHSATP | EDHCRRSWSS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DSTDSVISSE | SGNTYYRVVL | IGEQGVGKST | LANIFAGVHD | SMDSDCEVLG | EDTYERTLMV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DGESATIILL | DMWENKGENE | WLHDHCMQVG | DAYLIVYSIT | DRASFEKASE | LRIQLRRARQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TEDIPIILVG | NKSDLVRCRE | VSVSEGRACA | VVFDCKFIET | SAAVQHNVKE | LFEGIVRQVR |
| 250 | 260 | 270 | 280 | 290 | |
| LRRDSKEKNE | RRLAYQKRKE | SMPRKARRFW | GKIVAKNNKN | MAFKLKSKSC | HDLSVL |