P54761
Gene name |
Ephb4 (Htk, Mdk2, Myk1) |
Protein name |
Ephrin type-B receptor 4 |
Names |
Developmental kinase 2, mDK-2, Hepatoma transmembrane kinase, Tyrosine kinase MYK-1 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:13846 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with P29323)
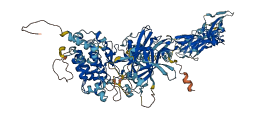
Ephrin type-B receptor 2 is a membrane-associated protein that mediates axon guidance, cell migration and morphogenesis. The Eph receptor tyrosine kinase family is regulated by autophosphorylation within the juxtamembrane region and the kinase activation segment. The structure, supported by mutagenesis data, reveals that the juxtamembrane segment adopts a helical conformation that distorts the small lobe of the kinase domain, and blocks the activation segment from attaining an activated conformation. Phosphorylation of the conserved juxtamembrane tyrosines would relieve this autoinhibition by disturbing the association of the juxtamembrane segment with the kinase domain, while liberating phosphotyrosine sites for binding SH2 domains of target proteins.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
757-785 (Activation loop from InterPro)
Target domain |
615-899 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for P54761
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P54761-F1 | Predicted | AlphaFoldDB |
46 variants for P54761
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388797763 | 42 | W>* | No | EVA | |
| rs3388800388 | 56 | R>C | No | EVA | |
| rs3388802473 | 67 | G>W | No | EVA | |
| rs3388770065 | 79 | P>Q | No | EVA | |
| rs3388770074 | 116 | Y>H | No | EVA | |
| rs1134098407 | 150 | R>C | No | EVA | |
| rs3396328533 | 181 | Q>EPCRLLGPGKPGRSQLC* | No | EVA | |
| rs256719724 | 229 | P>T | No | EVA | |
| rs215493603 | 232 | N>S | No | EVA | |
| rs3388800337 | 249 | Q>H | No | EVA | |
| rs3388770135 | 256 | A>S | No | EVA | |
| rs3388802777 | 267 | V>I | No | EVA | |
| rs3388797215 | 277 | K>N | No | EVA | |
| rs3388786671 | 316 | R>L | No | EVA | |
| rs3388800390 | 317 | S>T | No | EVA | |
| rs3388797780 | 338 | T>I | No | EVA | |
| rs3396326660 | 382 | G>* | No | EVA | |
| rs3388797772 | 384 | R>P | No | EVA | |
| rs3388806876 | 385 | D>H | No | EVA | |
| rs264512183 | 387 | V>I | No | EVA | |
| rs240038619 | 495 | R>Q | No | EVA | |
| rs3388802773 | 507 | V>L | No | EVA | |
| rs3388788909 | 554 | V>M | No | EVA | |
| rs3388800092 | 615 | V>I | No | EVA | |
| rs3388792238 | 632 | G>S | No | EVA | |
| rs1134499068 | 638 | G>V | No | EVA | |
| rs3388802511 | 664 | E>G | No | EVA | |
| rs3388770063 | 726 | S>P | No | EVA | |
| rs3388788926 | 781 | K>R | No | EVA | |
| rs3388781947 | 788 | A>V | No | EVA | |
| rs3396235336 | 791 | A>G | No | EVA | |
| rs3396283587 | 792 | I>N | No | EVA | |
| rs3396032079 | 793 | A>G | No | EVA | |
| rs3396192795 | 796 | K>E | No | EVA | |
| rs3388788929 | 798 | T>I | No | EVA | |
| rs3388788922 | 814 | M>I | No | EVA | |
| rs3388788912 | 830 | I>T | No | EVA | |
| rs3388802520 | 865 | P>S | No | EVA | |
| rs3388797149 | 876 | K>* | No | EVA | |
| rs3388788940 | 894 | A>T | No | EVA | |
| rs3388802476 | 898 | L>F | No | EVA | |
| rs3388797217 | 922 | G>R | No | EVA | |
| rs3388806917 | 967 | H>Y | No | EVA | |
| rs218420013 | 972 | A>T | No | EVA | |
| rs3388797730 | 976 | A>T | No | EVA | |
| rs230951901 | 982 | G>V | No | EVA |
No associated diseases with P54761
12 regional properties for P54761
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | WD40 repeat | 516 - 562 | IPR001680-1 |
| repeat | WD40 repeat | 565 - 610 | IPR001680-2 |
| repeat | WD40 repeat | 660 - 697 | IPR001680-3 |
| repeat | WD40 repeat | 703 - 742 | IPR001680-4 |
| repeat | WD40 repeat | 792 - 831 | IPR001680-5 |
| repeat | WD40 repeat | 832 - 869 | IPR001680-6 |
| repeat | WD40 repeat | 872 - 911 | IPR001680-7 |
| repeat | WD40 repeat | 918 - 956 | IPR001680-8 |
| repeat | WD40 repeat | 1032 - 1072 | IPR001680-9 |
| domain | Doublecortin domain | 166 - 253 | IPR003533-1 |
| domain | Doublecortin domain | 295 - 380 | IPR003533-2 |
| domain | HELP | 449 - 515 | IPR005108 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ephrin receptor activity | Combining with an ephrin receptor ligand to initiate a change in cell activity. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
| transmembrane-ephrin receptor activity | Combining with a transmembrane ephrin to initiate a change in cell activity. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| cell migration involved in sprouting angiogenesis | The orderly movement of endothelial cells into the extracellular matrix in order to form new blood vessels involved in sprouting angiogenesis. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| heart morphogenesis | The developmental process in which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. |
| positive regulation of aorta morphogenesis | Any process that activates or increases the frequency, rate or extent of aorta morphogenesis. |
| positive regulation of kinase activity | Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| positive regulation of T cell costimulation | Any process that activates or increases the frequency, rate or extent of T cell costimulation. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| venous blood vessel morphogenesis | The process in which the anatomical structures of venous blood vessels are generated and organized. Veins are blood vessels that transport blood from the body and its organs to the heart. |
86 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P29322 | EPHA8 | Ephrin type-A receptor 8 | Homo sapiens (Human) | SS |
| P21709 | EPHA1 | Ephrin type-A receptor 1 | Homo sapiens (Human) | SS |
| P54764 | EPHA4 | Ephrin type-A receptor 4 | Homo sapiens (Human) | SS |
| P54753 | EPHB3 | Ephrin type-B receptor 3 | Homo sapiens (Human) | SS |
| P29320 | EPHA3 | Ephrin type-A receptor 3 | Homo sapiens (Human) | PR |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| Q15375 | EPHA7 | Ephrin type-A receptor 7 | Homo sapiens (Human) | SS |
| Q5JZY3 | EPHA10 | Ephrin type-A receptor 10 | Homo sapiens (Human) | SS |
| P29323 | EPHB2 | Ephrin type-B receptor 2 | Homo sapiens (Human) | EV |
| Q9UF33 | EPHA6 | Ephrin type-A receptor 6 | Homo sapiens (Human) | SS |
| P54762 | EPHB1 | Ephrin type-B receptor 1 | Homo sapiens (Human) | SS |
| P54756 | EPHA5 | Ephrin type-A receptor 5 | Homo sapiens (Human) | SS |
| O15197 | EPHB6 | Ephrin type-B receptor 6 | Homo sapiens (Human) | SS |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| O08644 | Ephb6 | Ephrin type-B receptor 6 | Mus musculus (Mouse) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| Q00993 | Axl | Tyrosine-protein kinase receptor UFO | Mus musculus (Mouse) | PR |
| Q60805 | Mertk | Tyrosine-protein kinase Mer | Mus musculus (Mouse) | SS |
| Q9Z138 | Ror2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Mus musculus (Mouse) | SS |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| Q9Z139 | Ror1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Mus musculus (Mouse) | PR |
| P97793 | Alk | ALK tyrosine kinase receptor | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P35546 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Mus musculus (Mouse) | SS |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q62371 | Ddr2 | Discoidin domain-containing receptor 2 | Mus musculus (Mouse) | SS |
| Q03146 | Ddr1 | Epithelial discoidin domain-containing receptor 1 | Mus musculus (Mouse) | SS |
| P15209 | Ntrk2 | BDNF/NT-3 growth factors receptor | Mus musculus (Mouse) | SS |
| Q6VNS1 | Ntrk3 | NT-3 growth factor receptor | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35969 | Flt1 | Vascular endothelial growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| Q60751 | Igf1r | Insulin-like growth factor 1 receptor | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| P55144 | Tyro3 | Tyrosine-protein kinase receptor TYRO3 | Mus musculus (Mouse) | SS |
| P70424 | Erbb2 | Receptor tyrosine-protein kinase erbB-2 | Mus musculus (Mouse) | SS |
| Q61527 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Mus musculus (Mouse) | SS |
| Q61526 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Mus musculus (Mouse) | SS |
| Q01279 | Egfr | Epidermal growth factor receptor | Mus musculus (Mouse) | SS |
| Q01887 | Ryk | Tyrosine-protein kinase RYK | Mus musculus (Mouse) | PR |
| Q02858 | Tek | Angiopoietin-1 receptor | Mus musculus (Mouse) | SS |
| Q06806 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | Mus musculus (Mouse) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| O61460 | vab-1 | Ephrin receptor 1 | Caenorhabditis elegans | SS |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O13147 | ephb3 | Ephrin type-B receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73878 | ephb4b | Ephrin type-B receptor 4b | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MELRALLCWA | SLATALEETL | LNTKLETADL | KWVTYPQAEG | QWEELSGLDE | EQHSVRTYEV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CDMKRPGGQA | HWLRTGWVPR | RGAVHVYATI | RFTMMECLSL | PRASRSCKET | FTVFYYESEA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DTATAHTPAW | MENPYIKVDT | VAAEHLTRKR | PGAEATGKVN | IKTLRLGPLS | KAGFYLAFQD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QGACMALLSL | HLFYKKCSWL | ITNLTYFPET | VPRELVVPVA | GSCVANAVPT | ANPSPSLYCR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EDGQWAEQQV | TGCSCAPGYE | AAESNKVCRA | CGQGTFKPQI | GDESCLPCPA | NSHSNNIGSP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VCLCRIGYYR | ARSDPRSSPC | TTPPSAPRSV | VHHLNGSTLR | LEWSAPLESG | GREDLTYAVR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| CRECRPGGSC | LPCGGDMTFD | PGPRDLVEPW | VAIRGLRPDV | TYTFEVAALN | GVSTLATGPP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PFEPVNVTTD | REVPPAVSDI | RVTRSSPSSL | ILSWAIPRAP | SGAVLDYEVK | YHEKGAEGPS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SVRFLKTSEN | RAELRGLKRG | ASYLVQVRAR | SEAGYGPFGQ | EHHSQTQLDE | SESWREQLAL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| IAGTAVVGVV | LVLVVVIIAV | LCLRKQSNGR | EVEYSDKHGQ | YLIGHGTKVY | IDPFTYEDPN |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EAVREFAKEI | DVSYVKIEEV | IGAGEFGEVC | RGRLKAPGKK | ESCVAIKTLK | GGYTERQRRE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| FLSEASIMGQ | FEHPNIIRLE | GVVTNSVPVM | ILTEFMENGA | LDSFLRLNDG | QFTVIQLVGM |
| 730 | 740 | 750 | 760 | 770 | 780 |
| LRGIASGMRY | LAEMSYVHRD | LAARNILVNS | NLVCKVSDFG | LSRFLEENSS | DPTYTSSLGG |
| 790 | 800 | 810 | 820 | 830 | 840 |
| KIPIRWTAPE | AIAFRKFTSA | SDAWSYGIVM | WEVMSFGERP | YWDMSNQDVI | NAIEQDYRLP |
| 850 | 860 | 870 | 880 | 890 | 900 |
| PPPDCPTSLH | QLMLDCWQKD | RNARPRFPQV | VSALDKMIRN | PASLKIVARE | NGGASHPLLD |
| 910 | 920 | 930 | 940 | 950 | 960 |
| QRQPHYSAFG | SVGEWLRAIK | MGRYEESFAA | AGFGSFEVVS | QISAEDLLRI | GVTLAGHQKK |
| 970 | 980 | ||||
| ILASVQHMKS | QAKPGAPGGT | GGPAQQF |