P54646
Gene name |
PRKAA2 (AMPK, AMPK2) |
Protein name |
5'-AMP-activated protein kinase catalytic subunit alpha-2 |
Names |
AMPK subunit alpha-2, Acetyl-CoA carboxylase kinase, ACACA kinase, Hydroxymethylglutaryl-CoA reductase kinase, HMGCR kinase |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5563 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
MAP/MICROTUBULE AFFINITY-REGULATING KINASE (PTHR24346) |
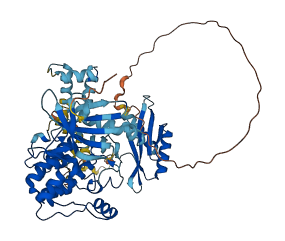
Descriptions
AMP-activated protein kinase (AMPK) acts as an energy sensor, being activated by metabolic stresses and regulating cellular metabolism. AMPK is a heterotrimer consisting of a catalytic alpha subunit and two regulatory subunits, beta and gamma. The human AMPK alpha2 kinase domain adopts the autoinhibited structure of the DFG-out conformation. The autoinhibitory domain following the kinase domain contributes to the stabilization of the DFG-out autoinhibitory structure. AMPK alpha2 activation is mediated by the phosphorylation of Thr located in the activation loop. The phosphorylated and/or unphosphorylated Thr interacts with residues in the regulatory subunits which stabilize the kinase-domain conformation in either the active or inactive mode.
Autoinhibitory domains (AIDs)
Target domain |
16-268 (Protein kinase domain) |
Relief mechanism |
PTM, Ligand binding |
Assay |
Structural analysis |
Accessory elements
156-178 (Activation loop from InterPro)
Target domain |
16-268 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Pang T et al. (2007) "Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits", The Journal of biological chemistry, 282, 495-506
- Hardie DG et al. (2012) "AMP-activated protein kinase: a target for drugs both ancient and modern", Chemistry & biology, 19, 1222-36
- Handa N et al. (2011) "Structural basis for compound C inhibition of the human AMP-activated protein kinase α2 subunit kinase domain", Acta crystallographica. Section D, Biological crystallography, 67, 480-7
- Littler DR et al. (2010) "A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation", Acta crystallographica. Section F, Structural biology and crystallization communications, 66, 143-51
Autoinhibited structure
Activated structure

14 structures for P54646
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2H6D | X-ray | 185 A | A | 6-279 | PDB |
| 2LTU | NMR | - | A | 282-339 | PDB |
| 2YZA | X-ray | 302 A | A | 6-279 | PDB |
| 3AQV | X-ray | 208 A | A | 6-279 | PDB |
| 4CFE | X-ray | 302 A | A/C | 1-552 | PDB |
| 4CFF | X-ray | 392 A | A/C | 1-552 | PDB |
| 4ZHX | X-ray | 299 A | A/C | 2-552 | PDB |
| 5EZV | X-ray | 299 A | PDB | ||
| 5ISO | X-ray | 263 A | A/C | 1-552 | PDB |
| 6B1U | X-ray | 277 A | A/C | 2-552 | PDB |
| 6B2E | X-ray | 380 A | A | 2-552 | PDB |
| 6BX6 | X-ray | 290 A | A | 6-279 | PDB |
| 7MYJ | X-ray | 295 A | A/C | 2-552 | PDB |
| AF-P54646-F1 | Predicted | AlphaFoldDB |
338 variants for P54646
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA340495205 rs1428540628 |
2 | A>V | No |
ClinGen TOPMed |
|
|
rs1480741512 CA340495250 |
5 | Q>E | No |
ClinGen gnomAD |
|
|
CA340495256 rs1177666094 |
5 | Q>R | No |
ClinGen gnomAD |
|
|
CA340495301 rs1196595450 |
7 | H>Q | No |
ClinGen TOPMed |
|
|
CA873655 rs200643979 |
8 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1235784068 CA340495307 |
8 | D>H | No |
ClinGen gnomAD |
|
|
rs1434366087 CA340495332 |
9 | G>E | No |
ClinGen TOPMed |
|
|
CA340495336 rs1434366087 |
9 | G>V | No |
ClinGen TOPMed |
|
|
rs752436954 CA873656 |
12 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873657 rs757985759 |
13 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs966678434 CA22805836 |
14 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs763759018 CA873658 |
15 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA340495421 rs1330973554 |
15 | H>Y | No |
ClinGen gnomAD |
|
|
CA873660 rs751060247 |
16 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA22805844 rs868402020 |
16 | Y>C | No |
ClinGen Ensembl |
|
|
CA340495463 rs1366799809 |
17 | V>G | No |
ClinGen gnomAD |
|
|
rs1304339883 CA340495451 |
17 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA873661 rs781236377 |
20 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340495563 rs1295726050 |
24 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA340495565 rs1295726050 |
24 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA22805886 rs867333017 |
25 | G>D | No |
ClinGen Ensembl |
|
|
rs745675639 CA873662 |
25 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA340495596 rs1236124861 |
26 | T>N | No |
ClinGen TOPMed |
|
|
CA340495614 rs749587981 |
27 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA873667 rs774427325 |
32 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1296094303 CA340502020 |
34 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
CA22820525 rs879141144 |
36 | Q>H | No |
ClinGen Ensembl |
|
|
CA22820530 rs1032540308 |
38 | T>P | No |
ClinGen TOPMed |
|
|
rs867367938 CA340502095 |
39 | G>C | No |
ClinGen Ensembl |
|
|
rs867367938 CA22820533 |
39 | G>S | No |
ClinGen Ensembl |
|
|
CA340502155 rs1448252942 |
44 | V>G | No |
ClinGen gnomAD |
|
|
rs1407331305 CA340502179 |
48 | N>D | No |
ClinGen TOPMed |
|
|
rs866247120 COSM1584641 CA22820542 COSM911062 |
53 | R>C | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
COSM1584640 CA873688 rs370153538 COSM911063 |
53 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA340502317 rs1224957749 |
58 | V>A | No |
ClinGen gnomAD |
|
|
CA22820558 rs978307258 |
61 | I>M | No |
ClinGen Ensembl |
|
|
rs748248170 CA873689 |
62 | K>I | No |
ClinGen ExAC gnomAD |
|
|
CA340502364 rs1284409982 |
62 | K>Q | No |
ClinGen gnomAD |
|
|
CA873690 rs373328629 |
63 | R>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1480644501 CA340502380 |
63 | R>Q | No |
ClinGen TOPMed |
|
|
CA873693 COSM1263042 COSM1263043 rs747610142 |
72 | R>C | oesophagus skin [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA873694 rs771285921 COSM1646202 COSM681676 |
72 | R>H | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1471971409 CA340502556 |
75 | H>L | No |
ClinGen gnomAD |
|
|
CA340488806 rs1396822539 |
80 | Y>C | No |
ClinGen gnomAD |
|
|
CA22792679 rs61765460 |
84 | S>I | No |
ClinGen Ensembl |
|
|
rs1340495433 CA340488842 |
85 | T>I | No |
ClinGen TOPMed |
|
|
CA873708 rs758607652 |
88 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA22792724 rs777923814 |
92 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA873710 rs777923814 |
92 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA873711 rs746995531 |
93 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1341618270 CA340488904 |
94 | E>G | No |
ClinGen gnomAD |
|
|
CA340488931 rs1385705903 |
98 | G>V | No |
ClinGen TOPMed |
|
|
rs1333320495 CA340488980 |
105 | I>F | No |
ClinGen gnomAD |
|
|
CA340488979 rs1333320495 |
105 | I>V | No |
ClinGen gnomAD |
|
|
CA340489018 rs746358536 |
110 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA873714 rs746358536 |
110 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA873713 rs781506978 |
110 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873733 rs780575560 |
111 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs906020143 CA22793389 |
112 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA22793392 rs906020143 |
112 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA873734 rs749593228 |
114 | M>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749593228 CA340489063 |
114 | M>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771787463 CA873735 |
115 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873737 rs746649362 |
117 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA873739 RCV000785689 rs145987132 |
118 | R>W | No |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
CA873741 rs765092888 |
119 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA340489154 rs1163120480 |
120 | F>S | No |
ClinGen gnomAD |
|
|
CA340489166 rs1175956445 |
121 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs762691181 CA873743 |
123 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs763930816 CA873744 |
124 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1315124138 CA340489319 |
130 | C>Y | No |
ClinGen gnomAD |
|
|
rs757499355 CA873746 |
137 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA340489452 rs1244151799 |
138 | R>* | No |
ClinGen gnomAD |
|
|
CA873747 rs190741217 |
138 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA873752 rs755346871 |
148 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1328701024 CA340489621 |
149 | A>V | No |
ClinGen gnomAD |
|
|
CA873754 rs17848596 |
150 | H>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA340489657 rs1422953446 |
153 | A>V | No |
ClinGen gnomAD |
|
|
rs770561050 CA873755 |
154 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs929309136 CA22793507 |
155 | I>V | No |
ClinGen TOPMed |
|
|
rs1457982338 CA340490510 |
159 | G>E | No |
ClinGen gnomAD |
|
|
CA340489693 COSM911071 COSM911070 rs902215574 |
159 | G>R | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs902215574 COSM911071 CA22793521 COSM911070 |
159 | G>R | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1234093391 CA340490521 |
161 | S>P | No |
ClinGen TOPMed |
|
|
rs780775307 CA873774 |
163 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA340490535 rs1306919952 |
163 | M>V | No |
ClinGen TOPMed |
|
|
rs1479931552 CA340490571 |
168 | E>K | No |
ClinGen gnomAD |
|
|
CA873776 rs371637215 |
175 | G>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA22794456 rs866103650 |
180 | A>V | No |
ClinGen Ensembl |
|
|
rs1407170093 CA340490722 |
188 | R>S | No |
ClinGen gnomAD |
|
|
rs1557560303 CA340490743 |
191 | A>V | No |
ClinGen Ensembl |
|
|
rs1323669301 CA340490775 |
196 | D>G | No |
ClinGen TOPMed |
|
|
CA340490778 rs1442773327 |
197 | I>V | No |
ClinGen TOPMed |
|
|
rs1349223592 CA340490810 |
201 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA340490825 rs1422713891 |
203 | I>N | No |
ClinGen gnomAD |
|
|
COSM1600268 CA873796 rs755752772 COSM1600267 |
207 | L>F | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs748762841 CA873799 |
211 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748762841 CA873798 |
211 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873800 rs151323778 |
217 | E>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA340490993 rs1325338179 |
218 | H>Q | No |
ClinGen gnomAD |
|
|
CA340490988 rs1569782596 |
218 | H>R | No |
ClinGen Ensembl |
|
|
CA340491005 rs1288534175 |
219 | V>A | No |
ClinGen gnomAD |
|
|
CA873801 rs748011810 |
221 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1473937464 CA340491081 |
224 | K>T | No |
ClinGen gnomAD |
|
|
rs773135709 CA873803 COSM166749 |
227 | R>* | large_intestine haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs761035255 CA873804 |
227 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873805 rs771213351 |
228 | G>W | No |
ClinGen ExAC gnomAD |
|
|
rs776607985 CA873806 |
229 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA340491213 rs1418880681 |
232 | Y>C | No |
ClinGen gnomAD |
|
|
rs1316031210 CA340491223 |
233 | I>V | No |
ClinGen gnomAD |
|
|
CA873807 rs759612307 |
236 | Y>D | No |
ClinGen ExAC |
|
|
rs765209932 CA873808 |
238 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs141766544 COSM3491097 CA22796043 COSM107875 |
239 | R>C | skin prostate [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA873809 rs755095917 COSM1738569 COSM1738570 |
239 | R>H | urinary_tract haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1448347506 CA340491317 |
240 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
rs763450384 CA873812 |
242 | A>D | No |
ClinGen ExAC gnomAD |
|
|
COSM265356 rs1031612391 CA22796065 |
242 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs369893630 CA873813 |
243 | T>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs369546032 CA22796084 |
247 | H>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA873816 rs534372739 |
250 | Q>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1239521525 CA340491384 |
251 | V>D | No |
ClinGen TOPMed gnomAD |
|
|
CA873817 rs753307011 |
251 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA873819 rs778254708 |
256 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM3865863 COSM3865862 CA340491441 rs1442861086 |
256 | R>Q | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs267598662 CA873820 |
258 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA22796149 rs985601596 |
259 | I>M | No |
ClinGen Ensembl |
|
|
rs758394696 CA873821 |
259 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA873822 rs777723104 |
260 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs746762993 CA873823 |
261 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs771303312 CA873824 |
262 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs776961274 CA873825 |
263 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA340492602 rs1398950938 |
264 | E>D | No |
ClinGen gnomAD |
|
|
rs1170749841 CA340492597 |
264 | E>Q | No |
ClinGen gnomAD |
|
|
rs1327443948 CA340492610 |
265 | H>R | No |
ClinGen gnomAD |
|
|
rs1391281720 CA340492606 |
265 | H>Y | No |
ClinGen gnomAD |
|
|
rs182000380 CA873843 |
266 | E>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs190412282 CA873842 |
266 | E>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs753420724 CA22801181 |
270 | Q>E | No |
ClinGen gnomAD |
|
|
rs1258735622 CA340492705 |
271 | D>G | No |
ClinGen gnomAD |
|
|
CA22801191 rs1017992313 |
272 | L>F | No |
ClinGen Ensembl |
|
|
rs1464949361 CA340492724 |
273 | P>A | No |
ClinGen TOPMed |
|
|
rs1338856347 CA340492752 |
275 | Y>H | No |
ClinGen gnomAD |
|
|
rs1193933257 CA340492790 |
277 | F>S | No |
ClinGen gnomAD |
|
|
CA873844 rs757116836 |
279 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1266576242 CA340492848 |
281 | P>L | No |
ClinGen gnomAD |
|
|
rs1197866775 CA340492845 |
281 | P>S | No |
ClinGen gnomAD |
|
|
rs1478571313 CA340492888 |
284 | D>A | No |
ClinGen gnomAD |
|
|
rs746162985 CA873846 |
285 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA873847 rs769852348 |
286 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA340492923 rs775647745 |
286 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM426507 rs963948073 COSM1474105 CA22801229 |
287 | V>I | pancreas breast [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs369053692 CA873849 |
289 | D>G | No |
ClinGen ESP ExAC gnomAD |
|
|
rs370838457 CA22801231 |
290 | D>H | No |
ClinGen ESP TOPMed |
|
|
rs370838457 CA340492970 |
290 | D>N | No |
ClinGen ESP TOPMed |
|
|
COSM1748524 COSM1748525 rs1302900503 CA340492983 |
291 | E>K | urinary_tract [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA340493015 rs77233967 |
293 | V>A | No |
ClinGen gnomAD |
|
|
CA22801241 rs77233967 |
293 | V>G | No |
ClinGen gnomAD |
|
|
CA22801237 rs78480732 |
293 | V>L | No |
ClinGen Ensembl |
|
|
CA340493034 rs1402031954 |
296 | V>L | No |
ClinGen gnomAD |
|
|
CA340493033 rs1402031954 |
296 | V>M | No |
ClinGen gnomAD |
|
|
CA873850 rs769171470 |
300 | F>C | No |
ClinGen ExAC gnomAD |
|
|
rs774884393 CA873851 |
302 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762273163 CA873852 |
303 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1262113124 CA340493172 |
305 | S>P | No |
ClinGen TOPMed |
|
|
rs1199822548 CA340493223 |
308 | M>I | No |
ClinGen TOPMed |
|
|
rs773435420 CA873854 |
311 | L>* | No |
ClinGen ExAC |
|
|
rs951220031 CA22801266 |
314 | G>C | No |
ClinGen Ensembl |
|
|
rs764728833 CA873856 |
320 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1489151374 CA340493383 |
323 | A>D | No |
ClinGen gnomAD |
|
|
rs1216813279 CA340493397 |
324 | Y>C | No |
ClinGen gnomAD |
|
|
rs866588635 CA22801300 |
325 | H>Y | No |
ClinGen Ensembl |
|
|
rs762602789 CA873858 |
327 | I>V | No |
ClinGen ExAC gnomAD |
|
|
COSM911079 CA873862 COSM911078 rs373885346 |
331 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA873861 rs763667032 |
331 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340493528 rs1473382427 |
334 | M>L | No |
ClinGen gnomAD |
|
|
CA873864 rs781056860 |
336 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs750122592 CA873865 |
338 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA340493611 rs1229080560 |
339 | E>A | No |
ClinGen TOPMed |
|
|
rs377396399 CA340493614 |
339 | E>D | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA340493630 rs1299267070 |
340 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1463086956 CA340493633 |
341 | Y>H | No |
ClinGen gnomAD |
|
|
CA340493650 rs1394632782 |
342 | L>F | No |
ClinGen TOPMed |
|
|
rs370738439 CA873867 |
343 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749485325 CA873868 |
344 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA22801345 rs749485325 |
344 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340493683 rs1368711403 |
345 | S>G | No |
ClinGen TOPMed |
|
|
rs1169896197 CA340493687 |
345 | S>N | No |
ClinGen TOPMed |
|
|
CA340493689 rs1169896197 |
345 | S>T | No |
ClinGen TOPMed |
|
|
CA873869 rs768760746 |
349 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1312849166 CA340493787 |
353 | D>G | No |
ClinGen gnomAD |
|
|
rs1435128495 CA340493807 |
354 | D>E | No |
ClinGen TOPMed |
|
|
CA340493797 rs1355840765 |
354 | D>Y | No |
ClinGen gnomAD |
|
|
CA873871 rs565908999 |
355 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA873872 rs141573879 |
356 | A>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1259606088 CA340493845 |
357 | M>I | No |
ClinGen TOPMed |
|
|
rs1201844736 CA340493859 |
358 | H>R | No |
ClinGen gnomAD |
|
|
CA340493879 rs1211439926 |
360 | P>S | No |
ClinGen TOPMed |
|
|
rs1256189019 CA340493893 |
361 | P>S | No |
ClinGen gnomAD |
|
|
CA340493904 rs1309387242 |
362 | G>D | No |
ClinGen gnomAD |
|
|
CA873873 rs376352128 |
362 | G>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1309387242 CA340493905 |
362 | G>V | No |
ClinGen gnomAD |
|
|
CA22801389 rs903921338 |
365 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA340493935 rs903921338 |
365 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA22801391 rs936769928 |
373 | L>F | No |
ClinGen TOPMed |
|
|
CA22801393 rs370525892 |
374 | I>T | No |
ClinGen gnomAD |
|
|
CA22801398 rs945537313 |
376 | D>G | No |
ClinGen TOPMed |
|
|
rs1159611433 CA340494086 |
377 | S>N | No |
ClinGen gnomAD |
|
|
CA340494104 rs1557564036 |
378 | P>L | No |
ClinGen Ensembl |
|
|
rs1353386070 CA340494114 |
379 | K>R | No |
ClinGen gnomAD |
|
|
CA340494136 rs1304813519 |
381 | R>K | No |
ClinGen gnomAD |
|
|
CA873876 rs775324119 |
383 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs571707957 CA873878 |
385 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA340494207 rs1240848958 |
386 | A>V | No |
ClinGen gnomAD |
|
|
CA873880 rs566846969 |
389 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA22801422 rs566846969 |
389 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1256237325 CA340494246 |
390 | T>A | No |
ClinGen gnomAD |
|
|
rs1557564126 CA340494281 |
393 | K>E | No |
ClinGen Ensembl |
|
|
CA340494301 rs1322462262 |
394 | S>C | No |
ClinGen gnomAD |
|
|
rs1322462262 CA340494302 |
394 | S>F | No |
ClinGen gnomAD |
|
|
rs1557564137 CA340494321 |
396 | A>P | No |
ClinGen Ensembl |
|
|
CA873882 rs750250924 |
397 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1008014097 CA22801439 |
400 | A>V | No |
ClinGen Ensembl |
|
|
rs1557564159 CA340494407 |
402 | W>* | No |
ClinGen Ensembl |
|
|
CA340494429 rs1409708472 |
405 | G>R | No |
ClinGen TOPMed |
|
|
CA873884 rs779791540 |
406 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs754122743 CA873886 |
407 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340494479 rs1250174544 |
409 | Q>* | No |
ClinGen gnomAD |
|
|
rs1331031283 CA340494500 |
410 | S>N | No |
ClinGen TOPMed |
|
|
CA22801457 rs370382985 |
410 | S>R | No |
ClinGen gnomAD |
|
|
rs557090880 CA873889 |
412 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA340494594 rs1557564224 |
416 | M>I | No |
ClinGen Ensembl |
|
|
CA340494579 rs1402575395 |
416 | M>V | No |
ClinGen gnomAD |
|
|
rs995187567 CA22801492 |
417 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs747519938 CA873892 |
420 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747519938 CA340494637 |
420 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873893 rs574007984 |
421 | R>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA873894 rs777124193 |
421 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762711078 CA873895 |
423 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA340494720 rs1006538361 |
426 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1318009672 CA340494747 |
428 | F>V | No |
ClinGen TOPMed gnomAD |
|
|
CA873908 rs752943476 |
432 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340494860 rs752943476 |
432 | V>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873909 rs758448281 |
433 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs747529167 CA873912 |
437 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA873913 rs200975038 |
438 | L>F | No |
ClinGen 1000Genomes ExAC |
|
|
CA340494901 rs1417490638 COSM171102 |
439 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA22802379 rs374865775 |
439 | R>H | No |
ClinGen 1000Genomes ESP TOPMed gnomAD |
|
|
rs374865775 CA340494905 |
439 | R>L | No |
ClinGen 1000Genomes ESP TOPMed gnomAD |
|
|
CA340494906 rs1164141817 |
440 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA340494908 rs1164141817 |
440 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA340494938 rs781694870 |
444 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873914 rs781694870 |
444 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873915 rs147304494 |
445 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA340494954 rs1458906904 |
447 | T>S | No |
ClinGen TOPMed |
|
|
rs767166675 CA340494961 |
448 | G>D | No |
ClinGen gnomAD |
|
|
rs900530381 CA22802394 |
448 | G>S | No |
ClinGen Ensembl |
|
|
CA22802395 rs767166675 |
448 | G>V | No |
ClinGen gnomAD |
|
|
rs773919981 CA873917 |
449 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs144733520 CA873919 |
451 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA340495002 rs1380749074 |
454 | S>N | No |
ClinGen gnomAD |
|
|
rs1230460047 CA340495017 |
456 | Q>P | No |
ClinGen gnomAD |
|
|
rs1230460047 CA340495018 |
456 | Q>R | No |
ClinGen gnomAD |
|
|
CA340495035 rs1359425212 |
458 | Y>* | No |
ClinGen TOPMed gnomAD |
|
|
CA22802436 rs60557166 |
459 | L>M | No |
ClinGen Ensembl |
|
|
rs372218435 CA873921 |
461 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA340495073 rs1288649641 |
464 | S>I | No |
ClinGen gnomAD |
|
|
CA22802452 rs868525835 |
471 | S>N | No |
ClinGen Ensembl |
|
|
CA873924 rs759277859 |
472 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1024487611 CA22802458 |
472 | I>V | No |
ClinGen TOPMed |
|
|
CA340495713 rs1557565624 |
478 | E>V | No |
ClinGen Ensembl |
|
|
CA340495718 rs1426820195 |
479 | Q>E | No |
ClinGen gnomAD |
|
|
rs1470029924 CA340495724 |
479 | Q>H | No |
ClinGen gnomAD |
|
|
CA340495753 rs1403308803 |
484 | S>* | No |
ClinGen gnomAD |
|
|
CA873944 rs374779642 COSM3944195 COSM3944196 |
484 | S>P | ovary [Cosmic] | No |
ClinGen cosmic curated ESP ExAC |
|
rs368958705 CA873947 |
485 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA340495765 rs1336935547 |
486 | P>R | No |
ClinGen gnomAD |
|
|
rs762036952 CA873948 |
487 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373434788 CA873949 |
487 | Q>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373434788 CA340495769 |
487 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA873951 rs756673522 |
488 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs142684565 CA22803175 |
488 | R>H | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA873952 rs780292097 |
489 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA873954 rs199623319 |
490 | C>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA873953 rs199623319 |
490 | C>Y | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA873955 rs777434133 |
491 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA340495811 rs1394547484 |
492 | A>T | No |
ClinGen TOPMed |
|
|
CA340495821 rs1437231136 |
493 | A>P | No |
ClinGen gnomAD |
|
|
CA340495823 rs1437231136 |
493 | A>S | No |
ClinGen gnomAD |
|
|
CA873956 rs746641545 |
494 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs746641545 CA340495840 |
494 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA340495848 rs1480530584 |
495 | L>* | No |
ClinGen gnomAD |
|
|
rs553841842 CA873958 |
498 | P>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs769707635 CA873960 |
500 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA340495894 rs1557565736 |
500 | S>L | No |
ClinGen Ensembl |
|
|
CA873962 rs762763141 |
501 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs775182044 CA873961 |
501 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1304428290 CA340495906 |
502 | F>C | No |
ClinGen TOPMed |
|
|
rs768318008 CA873963 |
503 | D>G | No |
ClinGen ExAC gnomAD |
|
|
COSM163722 COSM163721 CA340495910 rs1470118543 |
503 | D>H | breast [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs774757541 CA873964 |
504 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
CA340495928 rs1399478568 COSM72288 COSM72287 |
506 | T>A | ovary large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA340495932 rs1405705256 |
506 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA22803207 rs901520719 |
508 | E>K | No |
ClinGen TOPMed |
|
|
rs762118531 CA873965 |
509 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA340495951 rs1373191174 |
509 | S>T | No |
ClinGen TOPMed |
|
|
rs767760470 CA340495953 |
510 | H>N | No |
ClinGen ExAC gnomAD |
|
|
rs767760470 CA873966 |
510 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs750469075 CA873967 |
513 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA340495980 rs570406612 |
514 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA873968 rs570406612 |
514 | G>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA340495987 COSM192565 rs1338639598 COSM192566 |
515 | S>F | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs766867800 CA873969 |
516 | L>V | No |
ClinGen ExAC gnomAD |
|
|
COSM2194725 COSM2194726 rs1557565795 CA340496010 |
519 | S>F | pancreas [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs779142006 CA873974 |
522 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs543322076 CA340496036 |
523 | S>R | No |
ClinGen gnomAD |
|
|
rs756846399 CA873976 |
524 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA340496037 rs1281999558 |
524 | T>P | No |
ClinGen gnomAD |
|
|
rs1475352043 CA340496074 |
530 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs991680488 CA22803246 |
531 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs769645165 CA873979 |
532 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA873980 rs780092172 |
533 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1264984845 CA340496092 |
533 | G>V | No |
ClinGen TOPMed |
|
|
CA873981 rs749153323 |
535 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs575965814 CA22803256 |
536 | T>S | No |
ClinGen 1000Genomes |
|
|
rs1006002898 CA22803261 |
537 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA340496126 rs1325959714 |
538 | D>E | No |
ClinGen gnomAD |
|
|
CA873982 rs768558815 |
538 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs748392243 CA873984 |
542 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA873983 rs200621520 |
542 | M>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA340496172 rs1360164867 |
544 | A>D | No |
ClinGen gnomAD |
|
|
CA340496169 rs1313641358 |
544 | A>S | No |
ClinGen TOPMed |
|
|
rs772351857 CA873985 |
545 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA873986 rs146730697 |
548 | T>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs760757646 CA873987 |
549 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA340496200 rs766345818 |
549 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA873988 rs766345818 |
549 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs555212847 CA873990 |
552 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA340496216 rs1387624099 |
552 | R>H | No |
ClinGen gnomAD |
No associated diseases with P54646
6 regional properties for P54646
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 16 - 268 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 135 - 147 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 22 - 45 | IPR017441 |
| domain | PRKAA2, UBA-like autoinhibitory domain | 285 - 349 | IPR028783 |
| domain | AMPK, C-terminal adenylate sensor domain | 401 - 479 | IPR032270 |
| domain | 5'-AMP-activated protein kinase catalytic subunit alpha-2, C-terminal | 395 - 550 | IPR039148 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24346 | MAP/MICROTUBULE AFFINITY-REGULATING KINASE |
| PANTHER Subfamily | PTHR24346:SF82 | SERINE_THREONINE-PROTEIN KINASE MARK-A-RELATED |
| PANTHER Protein Class |
non-receptor serine/threonine protein kinase
protein modifying enzyme |
|
| PANTHER Pathway Category |
p53 pathway by glucose deprivation AMPK |
|
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cytoplasmic stress granule | A dense aggregation in the cytosol composed of proteins and RNAs that appear when the cell is under stress. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nuclear speck | A discrete extra-nucleolar subnuclear domain, 20-50 in number, in which splicing factors are seen to be localized by immunofluorescence microscopy. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleotide-activated protein kinase complex | A protein complex that possesses nucleotide-dependent protein kinase activity. The nucleotide can be AMP (in S. pombe and human) or ADP (in S. cerevisiae). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
11 GO annotations of molecular function
| Name | Definition |
|---|---|
| [acetyl-CoA carboxylase] kinase activity | Catalysis of the reaction: ATP |
| [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity | +Catalysis of the reaction: [3-hydroxy-3-methylglutaryl-CoA reductase (NADPH)] + ATP = [3-hydroxy-3-methylglutaryl-CoA reductase (NADPH)] phosphate + ADP. |
| AMP-activated protein kinase activity | Catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. This reaction requires the presence of AMP. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| histone serine kinase activity | Catalysis of the transfer of a phosphate group to a serine residue of a histone. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
34 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cellular response to calcium ion | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a calcium ion stimulus. |
| cellular response to glucose starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of glucose. |
| cellular response to glucose stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| cellular response to nutrient levels | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients. |
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| cellular response to prostaglandin E stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a prostagladin E stimulus. |
| cellular response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organism exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| cholesterol biosynthetic process | The chemical reactions and pathways resulting in the formation of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. |
| chromatin organization | The assembly or remodeling of chromatin composed of DNA complexed with histones, other associated proteins, and sometimes RNA. |
| energy homeostasis | Any process involved in the balance between food intake (energy input) and energy expenditure. |
| fatty acid biosynthetic process | The chemical reactions and pathways resulting in the formation of a fatty acid, any of the aliphatic monocarboxylic acids that can be liberated by hydrolysis from naturally occurring fats and oils. Fatty acids are predominantly straight-chain acids of 4 to 24 carbon atoms, which may be saturated or unsaturated; branched fatty acids and hydroxy fatty acids also occur, and very long chain acids of over 30 carbons are found in waxes. |
| fatty acid homeostasis | Any process involved in the maintenance of an internal steady state of fatty acid within an organism or cell. |
| glucose homeostasis | Any process involved in the maintenance of an internal steady state of glucose within an organism or cell. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| lipid biosynthetic process | The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of TOR signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of TOR signaling. |
| negative regulation of tubulin deacetylation | Any process that stops, prevents or reduces the frequency, rate or extent of tubulin deacetylation. |
| positive regulation of autophagy | Any process that activates, maintains or increases the rate of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| positive regulation of glycolytic process | Any process that activates or increases the frequency, rate or extent of glycolysis. |
| positive regulation of macroautophagy | Any process, such as recognition of nutrient depletion, that activates or increases the rate of macroautophagy to bring cytosolic macromolecules to the vacuole/lysosome for degradation. |
| positive regulation of peptidyl-lysine acetylation | Any process that activates or increases the frequency, rate or extent of peptidyl-lysine acetylation. |
| positive regulation of protein localization | Any process that activates or increases the frequency, rate or extent of a protein localization. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of circadian rhythm | Any process that modulates the frequency, rate or extent of a circadian rhythm. A circadian rhythm is a biological process in an organism that recurs with a regularity of approximately 24 hours. |
| regulation of macroautophagy | Any process that modulates the frequency, rate or extent of macroautophagy. |
| regulation of microtubule cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins. |
| regulation of stress granule assembly | Any process that modulates the rate, frequency or extent of stress granule assembly, the aggregation, arrangement and bonding together of proteins and RNA molecules to form a stress granule. |
| response to muscle activity | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a muscle activity stimulus. |
| rhythmic process | Any process pertinent to the generation and maintenance of rhythms in the physiology of an organism. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
71 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q02066 | Abscisic acid-inducible protein kinase | Triticum aestivum (Wheat) | PR | |
| P06782 | SNF1 | Carbon catabolite-derepressing protein kinase | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | EV |
| Q5GLH2 | TRIB2 | Tribbles homolog 2 | Bos taurus (Bovine) | SS |
| Q0VCE3 | TRIB3 | Tribbles homolog 3 | Bos taurus (Bovine) | SS |
| Q3SZW1 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Bos taurus (Bovine) | PR |
| Q92519 | TRIB2 | Tribbles homolog 2 | Homo sapiens (Human) | SS |
| Q96RU7 | TRIB3 | Tribbles homolog 3 | Homo sapiens (Human) | PR |
| Q96RU8 | TRIB1 | Tribbles homolog 1 | Homo sapiens (Human) | EV |
| Q15831 | STK11 | Serine/threonine-protein kinase STK11 | Homo sapiens (Human) | PR |
| Q96RG2 | PASK | PAS domain-containing serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q9BXA7 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Homo sapiens (Human) | PR |
| O14757 | CHEK1 | Serine/threonine-protein kinase Chk1 | Homo sapiens (Human) | EV |
| Q8IWQ3 | BRSK2 | Serine/threonine-protein kinase BRSK2 | Homo sapiens (Human) | PR |
| Q8TDC3 | BRSK1 | Serine/threonine-protein kinase BRSK1 | Homo sapiens (Human) | SS |
| Q14680 | MELK | Maternal embryonic leucine zipper kinase | Homo sapiens (Human) | EV |
| P57059 | SIK1 | Serine/threonine-protein kinase SIK1 | Homo sapiens (Human) | PR |
| Q9NRH2 | SNRK | SNF-related serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| O60285 | NUAK1 | NUAK family SNF1-like kinase 1 | Homo sapiens (Human) | PR |
| Q13131 | PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Homo sapiens (Human) | EV |
| Q8K4K3 | Trib2 | Tribbles homolog 2 | Mus musculus (Mouse) | SS |
| Q61241 | Tssk1b | Testis-specific serine/threonine-protein kinase 1 | Mus musculus (Mouse) | PR |
| Q8K4K2 | Trib3 | Tribbles homolog 3 | Mus musculus (Mouse) | SS |
| O54863 | Tssk2 | Testis-specific serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q8K4K4 | Trib1 | Tribbles homolog 1 | Mus musculus (Mouse) | SS |
| Q5EG47 | Prkaa1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Mus musculus (Mouse) | SS |
| Q61846 | Melk | Maternal embryonic leucine zipper kinase | Mus musculus (Mouse) | PR |
| Q8BRK8 | Prkaa2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Mus musculus (Mouse) | SS |
| Q28948 | PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Sus scrofa (Pig) | SS |
| Q9WTQ6 | Trib3 | Tribbles homolog 3 | Rattus norvegicus (Rat) | SS |
| P54645 | Prkaa1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Rattus norvegicus (Rat) | EV |
| Q09137 | Prkaa2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Rattus norvegicus (Rat) | EV |
| Q5QNM6 | CIPK13 | Putative CBL-interacting protein kinase 13 | Oryza sativa subsp japonica (Rice) | PR |
| Q5JLQ9 | CIPK30 | CBL-interacting protein kinase 30 | Oryza sativa subsp japonica (Rice) | PR |
| Q5N942 | SAPK4 | Serine/threonine-protein kinase SAPK4 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q0 | OSK3 | Serine/threonine protein kinase OSK3 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75LR7 | SAPK1 | Serine/threonine-protein kinase SAPK1 | Oryza sativa subsp japonica (Rice) | PR |
| Q75H77 | SAPK10 | Serine/threonine-protein kinase SAPK10 | Oryza sativa subsp japonica (Rice) | PR |
| Q7Y0B9 | SAPK8 | Serine/threonine-protein kinase SAPK8 | Oryza sativa subsp japonica (Rice) | PR |
| Q7XQP4 | SAPK7 | Serine/threonine-protein kinase SAPK7 | Oryza sativa subsp japonica (Rice) | PR |
| Q0D4J7 | SAPK2 | Serine/threonine-protein kinase SAPK2 | Oryza sativa subsp japonica (Rice) | PR |
| Q6ERS4 | CIPK16 | CBL-interacting protein kinase 16 | Oryza sativa subsp japonica (Rice) | PR |
| P0C5D6 | SAPK3 | Serine/threonine-protein kinase SAPK3 | Oryza sativa subsp japonica (Rice) | PR |
| Q75V57 | SAPK9 | Serine/threonine-protein kinase SAPK9 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q1 | OSK4 | Serine/threonine protein kinase OSK4 | Oryza sativa subsp. japonica (Rice) | SS |
| Q852Q2 | OSK1 | Serine/threonine protein kinase OSK1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LIG4 | CIPK3 | CBL-interacting protein kinase 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q6ZLP5 | CIPK23 | CBL-interacting protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAX3 | CIPK33 | CBL-interacting protein kinase 33 | Oryza sativa subsp japonica (Rice) | PR |
| Q2QY53 | CIPK32 | CBL-interacting protein kinase 32 | Oryza sativa subsp japonica (Rice) | PR |
| Q21017 | kin-29 | Serine/threonine-protein kinase kin-29 | Caenorhabditis elegans | SS |
| Q95ZQ4 | aak-2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Caenorhabditis elegans | SS |
| P45894 | aak-1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Caenorhabditis elegans | SS |
| P43291 | SRK2A | Serine/threonine-protein kinase SRK2A | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C958 | SRK2B | Serine/threonine-protein kinase SRK2B | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M9E9 | SRK2C | Serine/threonine-protein kinase SRK2C | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64812 | SRK2J | Serine/threonine-protein kinase SRK2J | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2V452 | CIPK3 | CBL-interacting serine/threonine-protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22971 | CIPK13 | CBL-interacting serine/threonine-protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39192 | SRK2D | Serine/threonine-protein kinase SRK2D | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O65554 | CIPK6 | CBL-interacting serine/threonine-protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q940H6 | SRK2E | Serine/threonine-protein kinase SRK2E | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43292 | SRK2G | Serine/threonine-protein kinase SRK2G | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FJ54 | CIPK20 | CBL-interacting serine/threonine-protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FFP9 | SRK2H | Serine/threonine-protein kinase SRK2H | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39193 | SRK2I | Serine/threonine-protein kinase SRK2I | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93VD3 | CIPK23 | CBL-interacting serine/threonine-protein kinase 23 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94CG0 | CIPK21 | CBL-interacting serine/threonine-protein kinase 21 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLZ3 | KIN12 | SNF1-related protein kinase catalytic subunit alpha KIN12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LDI3 | CIPK24 | CBL-interacting serine/threonine-protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P92958 | KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEKQKHDGR | VKIGHYVLGD | TLGVGTFGKV | KIGEHQLTGH | KVAVKILNRQ | KIRSLDVVGK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IKREIQNLKL | FRHPHIIKLY | QVISTPTDFF | MVMEYVSGGE | LFDYICKHGR | VEEMEARRLF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QQILSAVDYC | HRHMVVHRDL | KPENVLLDAH | MNAKIADFGL | SNMMSDGEFL | RTSCGSPNYA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| APEVISGRLY | AGPEVDIWSC | GVILYALLCG | TLPFDDEHVP | TLFKKIRGGV | FYIPEYLNRS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VATLLMHMLQ | VDPLKRATIK | DIREHEWFKQ | DLPSYLFPED | PSYDANVIDD | EAVKEVCEKF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ECTESEVMNS | LYSGDPQDQL | AVAYHLIIDN | RRIMNQASEF | YLASSPPSGS | FMDDSAMHIP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PGLKPHPERM | PPLIADSPKA | RCPLDALNTT | KPKSLAVKKA | KWHLGIRSQS | KPYDIMAEVY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RAMKQLDFEW | KVVNAYHLRV | RRKNPVTGNY | VKMSLQLYLV | DNRSYLLDFK | SIDDEVVEQR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SGSSTPQRSC | SAAGLHRPRS | SFDSTTAESH | SLSGSLTGSL | TGSTLSSVSP | RLGSHTMDFF |
| 550 | |||||
| EMCASLITTL | AR |