P51946
Gene name |
CCNH |
Protein name |
Cyclin-H |
Names |
MO15-associated protein, p34, p37 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:902 |
EC number |
|
Protein Class |
|
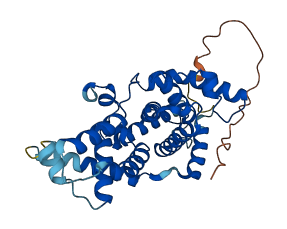
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

21 structures for P51946
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1JKW | X-ray | 260 A | A | 1-323 | PDB |
| 1KXU | X-ray | 260 A | A | 1-323 | PDB |
| 6O9L | EM | 720 A | 9 | 1-323 | PDB |
| 6XBZ | EM | 280 A | I | 1-323 | PDB |
| 6XD3 | EM | 330 A | I | 1-323 | PDB |
| 7B5O | EM | 250 A | I | 1-323 | PDB |
| 7B5Q | EM | 250 A | I | 1-323 | PDB |
| 7EGB | EM | 330 A | 9 | 1-323 | PDB |
| 7EGC | EM | 390 A | 9 | 1-323 | PDB |
| 7ENA | EM | 407 A | 9 | 1-323 | PDB |
| 7ENC | EM | 413 A | 9 | 1-323 | PDB |
| 7LBM | EM | 480 A | f | 1-323 | PDB |
| 7NVR | EM | 450 A | 9 | 1-323 | PDB |
| 8BVW | EM | 400 A | 9 | 1-323 | PDB |
| 8BYQ | EM | 410 A | 9 | 1-323 | PDB |
| 8GXQ | EM | 504 A | HJ | 1-323 | PDB |
| 8GXS | EM | 416 A | HJ | 1-323 | PDB |
| 8ORM | EM | 190 A | I | 1-323 | PDB |
| 8P6V | EM | 190 A | I | 1-323 | PDB |
| 8P6Y | EM | 190 A | I | 1-323 | PDB |
| AF-P51946-F1 | Predicted | AlphaFoldDB |
267 variants for P51946
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1249853544 CA360416557 |
2 | Y>F | No |
ClinGen gnomAD |
|
|
CA3336628 rs769622277 |
4 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA360416544 rs1213228300 |
4 | N>Y | No |
ClinGen gnomAD |
|
|
rs1353110518 CA360416538 |
5 | S>G | No |
ClinGen TOPMed |
|
|
CA360416531 rs1294230787 |
6 | S>G | No |
ClinGen TOPMed |
|
|
rs780883922 CA3336626 |
9 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA360416497 rs1353452063 |
10 | H>Q | No |
ClinGen gnomAD |
|
|
CA360416499 rs1489936872 |
10 | H>R | No |
ClinGen TOPMed |
|
|
rs1280709511 CA360416494 |
11 | W>R | No |
ClinGen gnomAD |
|
|
rs756919605 CA3336625 |
12 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA360416485 rs756919605 |
12 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs777180854 CA3336623 |
13 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA3336622 rs757878059 |
15 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1209019192 CA360416467 |
15 | S>R | No |
ClinGen TOPMed |
|
|
rs753194402 CA3336621 |
15 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs765742431 CA3336620 |
17 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs755881555 CA122464044 |
18 | Q>E | No |
ClinGen Ensembl |
|
|
CA3336618 rs755460097 |
20 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA360416430 rs1190157470 |
21 | R>G | No |
ClinGen gnomAD |
|
|
rs1561361246 CA360416428 |
21 | R>T | No |
ClinGen Ensembl |
|
|
CA360416421 rs1257146987 |
22 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
CA360416419 rs1257146987 |
22 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
CA360416422 rs1475623999 |
22 | L>V | No |
ClinGen gnomAD |
|
|
rs1036634923 CA122464043 |
23 | R>Q | No |
ClinGen gnomAD |
|
|
rs766790791 CA3336616 |
26 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760979973 CA3336615 |
27 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA122464041 rs995740892 |
28 | R>C | No |
ClinGen TOPMed |
|
|
CA3336614 rs2234942 |
28 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3336613 rs2234942 RCV000887401 VAR_013067 |
28 | R>L | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1283311173 CA360416355 |
32 | C>W | No |
ClinGen TOPMed gnomAD |
|
|
CA3336611 rs775360258 |
35 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1355684608 CA360416335 |
36 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 36 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs140234729 CA3336610 |
37 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
rs745750733 CA3336609 |
38 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1445499669 CA360416313 |
39 | K>R | No |
ClinGen gnomAD |
|
|
rs764097625 CA3336589 |
40 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs1194859349 CA360416279 |
42 | P>L | No |
ClinGen gnomAD |
|
|
rs760488477 CA3336585 |
43 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs772807882 CA3336584 |
44 | D>V | No |
ClinGen ExAC gnomAD |
|
|
CA3336582 rs747696109 |
46 | V>F | No |
ClinGen ExAC gnomAD |
|
|
CA3336583 rs747696109 |
46 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1227352914 CA360416244 |
48 | L>F | No |
ClinGen gnomAD |
|
|
CA122463899 rs922162838 |
49 | E>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA3336578 rs780679974 |
54 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs201537925 CA3336579 |
54 | M>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
VAR_013068 rs3093785 RCV000971510 CA3336580 |
54 | M>V | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1291728889 CA360416189 |
56 | L>F | No |
ClinGen TOPMed |
|
|
rs988750175 CA122463897 |
56 | L>H | No |
ClinGen Ensembl |
|
|
rs756471974 CA3336577 |
59 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs540142438 CA3336576 |
60 | Y>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs540142438 CA3336575 |
60 | Y>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1397219825 CA360416154 |
61 | E>* | No |
ClinGen TOPMed gnomAD |
|
|
CA360416156 rs1397219825 |
61 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs757507676 CA3336574 |
62 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1433055269 CA360416145 |
62 | K>R | No |
ClinGen gnomAD |
|
|
CA360416139 rs751743850 |
63 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336573 rs751743850 |
63 | R>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1423751655 CA360416140 |
63 | R>W | No |
ClinGen gnomAD |
|
|
CA3336571 rs763000210 |
66 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA122463896 rs1033044161 |
67 | F>L | No |
ClinGen Ensembl |
|
|
rs753813156 CA3336570 |
67 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1481884598 CA360416099 |
69 | S>A | No |
ClinGen gnomAD |
|
|
rs1282114823 COSM197634 CA360416095 |
69 | S>L | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs771696988 CA3336566 |
75 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1580469569 CA360416057 |
75 | M>T | No |
ClinGen Ensembl |
|
|
rs773071401 CA3336567 |
75 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs967855042 CA122463894 |
76 | P>L | No |
ClinGen gnomAD |
|
|
rs1283959344 CA360416048 |
77 | R>G | No |
ClinGen TOPMed |
|
|
rs1322704311 CA360416042 |
77 | R>S | No |
ClinGen TOPMed |
|
|
rs761557612 CA3336565 |
78 | S>F | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 78 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1224356231 CA360416029 |
80 | V>L | No |
ClinGen TOPMed |
|
|
rs936483643 CA122463717 |
81 | G>D | No |
ClinGen TOPMed |
|
|
rs1250379207 CA360416007 |
82 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA3336543 rs761675040 |
82 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs923725080 CA122463715 |
83 | A>S | No |
ClinGen TOPMed |
|
|
rs78508836 CA3336540 |
84 | C>F | No |
ClinGen ExAC |
|
|
CA360415996 rs1282922040 |
84 | C>R | No |
ClinGen TOPMed |
|
|
CA3336537 CA3336536 rs771368195 CA3336535 |
85 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs769150346 CA3336538 |
85 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA360415989 rs1484956549 |
85 | M>T | No |
ClinGen TOPMed |
|
|
rs769150346 CA3336539 |
85 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA3336533 rs778029154 |
86 | Y>F | No |
ClinGen ExAC |
|
|
rs747372568 CA3336534 |
86 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA3336532 rs377066266 |
87 | F>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs191853622 CA3336530 |
87 | F>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA360415970 rs1187877557 |
88 | K>R | No |
ClinGen gnomAD |
|
|
rs750420341 CA3336528 |
89 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs766815417 CA122463714 |
89 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs750420341 CA3336527 |
89 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762114015 CA3336524 |
91 | Y>* | No |
ClinGen ExAC TOPMed |
|
|
CA3336525 rs757182155 |
91 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA360415948 rs1267501058 |
92 | L>I | No |
ClinGen gnomAD |
|
| TCGA novel | 93 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3336523 rs751383225 |
95 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA360415900 rs146868495 |
98 | E>D | No |
ClinGen 1000Genomes ESP TOPMed gnomAD |
|
|
rs1244281017 CA360415883 |
100 | H>Q | No |
ClinGen gnomAD |
|
|
CA3336520 rs200476507 |
103 | I>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs781245392 CA3336504 |
105 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1385799292 CA360415835 |
106 | L>F | No |
ClinGen gnomAD |
|
|
CA360415830 rs1273518722 |
107 | T>A | No |
ClinGen gnomAD |
|
|
rs751505567 CA3336502 |
108 | C>F | No |
ClinGen ExAC gnomAD |
|
|
CA122463615 rs981585820 |
108 | C>W | No |
ClinGen Ensembl |
|
|
rs376356637 CA3336501 |
109 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1071041 rs758248932 CA3336500 |
110 | F>S | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1455486397 CA360415801 |
111 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1378187320 CA360415800 |
112 | A>T | No |
ClinGen gnomAD |
|
|
rs752434893 CA3336499 |
112 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA360415788 rs1449982012 |
113 | C>W | No |
ClinGen gnomAD |
|
| TCGA novel | 114 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA122463614 rs539013530 |
114 | K>T | No |
ClinGen 1000Genomes gnomAD |
|
|
CA360415774 rs1326598701 |
116 | D>N | No |
ClinGen gnomAD |
|
|
rs926007959 CA122463612 |
118 | F>S | No |
ClinGen TOPMed |
|
|
CA3336498 rs764792458 |
120 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA122463611 rs977456853 |
122 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA122463610 rs918830877 |
122 | S>N | No |
ClinGen TOPMed |
|
|
CA3336496 rs753302898 CA360415728 |
122 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336494 rs765806862 |
123 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs992035611 CA122463609 |
124 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA3336493 rs761199502 |
126 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs960592124 CA122463608 |
127 | G>E | No |
ClinGen TOPMed |
|
|
rs568535217 COSM1439022 CA3336492 |
127 | G>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC gnomAD |
|
rs143938518 CA3336491 |
128 | N>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1002033239 CA122463607 |
129 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs774697142 CA3336489 |
130 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336490 rs529200185 |
130 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA360415674 rs1171980817 |
132 | S>G | No |
ClinGen gnomAD |
|
|
CA360415662 rs1477015093 |
133 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA3336488 rs374176346 |
134 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749465599 CA3336487 |
134 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA360415658 rs749465599 |
134 | L>R | No |
ClinGen ExAC gnomAD |
|
|
COSM1200163 CA3336485 rs769947485 |
136 | Q>* | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA360415648 rs1213425729 |
136 | Q>R | No |
ClinGen gnomAD |
|
|
RCV000946861 CA3336484 VAR_013069 rs2266691 |
138 | K>R | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs756715232 CA122463605 |
139 | A>S | No |
ClinGen Ensembl |
|
|
rs777825509 CA3336483 |
139 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA360415621 rs1199203558 |
140 | L>P | No |
ClinGen gnomAD |
|
|
CA3336482 rs758336722 |
142 | Q>R | No |
ClinGen ExAC |
|
|
rs1269412442 CA360415581 |
146 | Y>C | No |
ClinGen gnomAD |
|
|
rs1353471497 CA360415584 |
146 | Y>N | No |
ClinGen gnomAD |
|
|
rs369461739 CA3336481 |
147 | E>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs778714787 CA3336480 |
151 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1180776806 CA360415551 |
151 | I>V | No |
ClinGen TOPMed |
|
|
rs1449930690 CA360395086 |
152 | Q>R | No |
ClinGen gnomAD |
|
| TCGA novel | 155 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3336479 rs199815978 |
155 | N>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1333302279 CA360395021 |
158 | L>H | No |
ClinGen gnomAD |
|
|
CA360395018 rs1412620316 |
159 | I>V | No |
ClinGen gnomAD |
|
|
CA3336477 rs765894978 |
161 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs1410002605 CA360394972 |
163 | P>L | No |
ClinGen gnomAD |
|
|
rs760155207 CA3336476 |
163 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA121832069 rs200767477 |
164 | Y>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3336475 rs200767477 |
164 | Y>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs769027195 CA3336471 |
169 | G>S | No |
ClinGen ExAC |
|
|
rs770037305 CA3336468 |
173 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1265725854 CA360394857 |
174 | L>I | No |
ClinGen gnomAD |
|
|
rs1226369619 CA360394715 |
176 | T>A | No |
ClinGen gnomAD |
|
|
COSM1695955 rs918052066 CA121831469 |
177 | R>C | skin [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA3336453 rs375955863 |
177 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3336452 rs201911431 |
178 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA121831466 rs201911431 |
178 | Y>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1437360102 CA360394619 |
185 | E>Q | No |
ClinGen gnomAD |
|
|
CA3336450 rs765542606 |
189 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA360394554 rs1580456311 |
191 | A>G | No |
ClinGen Ensembl |
|
|
CA360394557 rs1321887445 |
191 | A>S | No |
ClinGen gnomAD |
|
|
CA360394532 rs1431889359 |
194 | F>S | No |
ClinGen gnomAD |
|
|
rs759774462 CA3336449 |
196 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1448141189 COSM1311371 CA360394511 |
197 | R>K | Variant assessed as Somatic; 0.0 impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA3336448 rs145378845 COSM1071040 |
201 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs1483834494 CA360394444 |
203 | A>V | No |
ClinGen gnomAD |
|
|
rs771004517 CA3336447 |
204 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1360467420 CA360394428 |
205 | L>F | No |
ClinGen TOPMed |
|
|
CA3336445 rs138552646 |
207 | Y>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1343771894 CA360394396 |
208 | T>A | No |
ClinGen TOPMed |
|
|
rs1343771894 CA360394397 |
208 | T>P | No |
ClinGen TOPMed |
|
|
rs768375371 CA3336444 |
209 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs779736425 CA3336442 |
211 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA3336441 rs150007235 |
212 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3336440 rs778474752 |
214 | L>P | No |
ClinGen ExAC gnomAD |
|
| rs1307794299 | 215 | T>P | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756773005 CA3336439 |
219 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756773005 CA3336438 |
219 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs113599890 CA121831436 |
220 | S>G | No |
ClinGen Ensembl |
|
|
CA3336436 rs764661724 |
221 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA121831427 rs952174214 |
225 | G>E | No |
ClinGen Ensembl |
|
|
CA360394237 rs1285131522 |
225 | G>R | No |
ClinGen TOPMed |
|
|
CA360394228 rs1360115105 |
226 | I>V | No |
ClinGen TOPMed |
|
|
CA121831426 rs1029073278 |
227 | T>S | No |
ClinGen TOPMed |
|
|
rs1196667743 CA360394189 |
229 | E>G | No |
ClinGen gnomAD |
|
|
CA360394180 rs1473360844 |
230 | S>G | No |
ClinGen gnomAD |
|
|
rs1170103731 CA360393312 |
230 | S>R | No |
ClinGen TOPMed |
|
|
rs1323744504 CA360393250 COSM1544391 |
234 | E>D | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 237 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1377575843 CA360393218 |
237 | M>K | No |
ClinGen gnomAD |
|
|
CA3336407 rs766618619 |
242 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs760723804 CA3336406 |
243 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1334879201 CA360393152 |
243 | T>S | No |
ClinGen TOPMed |
|
|
rs750525738 CA3336405 |
244 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767426565 CA3336404 |
245 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA3336402 rs775386053 |
246 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336401 rs200152471 |
247 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs759325211 CA3336400 |
249 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA3336399 rs753326332 |
250 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA360393078 rs1376139691 |
250 | D>V | No |
ClinGen gnomAD |
|
|
rs770630511 CA3336398 |
251 | I>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA360393068 rs770630511 |
251 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336378 rs371630075 |
254 | S>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA360392716 rs1253903016 |
256 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 257 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA360392703 rs1206391717 |
257 | N>I | No |
ClinGen gnomAD |
|
|
rs1580445461 CA360392697 |
258 | L>V | No |
ClinGen Ensembl |
|
|
CA360392649 rs1268977589 |
260 | K>N | No |
ClinGen gnomAD |
|
|
CA360392636 rs1411103655 |
261 | K>R | No |
ClinGen TOPMed |
|
|
rs1301401427 CA360392612 |
262 | Y>* | No |
ClinGen gnomAD |
|
|
rs1461938466 CA360392562 |
265 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
VAR_013070 CA3336375 rs2230641 |
270 | V>A | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA121829933 rs2230641 |
270 | V>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1164479087 CA360392467 |
270 | V>I | No |
ClinGen TOPMed |
|
| TCGA novel | 271 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3336374 rs766049330 |
273 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs368654074 CA3336373 |
274 | K>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA360392409 rs1369850555 |
275 | Q>H | No |
ClinGen gnomAD |
|
|
CA121829923 rs972792143 |
275 | Q>R | No |
ClinGen Ensembl |
|
|
rs1166038228 CA360392397 |
276 | K>N | No |
ClinGen gnomAD |
|
|
rs771746887 CA3336371 |
277 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3336370 rs747656250 |
278 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs775055673 CA3336369 COSM3780817 |
279 | R>* | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA3336368 rs769299865 |
279 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749686780 CA360392353 |
280 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200031964 CA3336365 |
283 | A>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs200031964 CA3336366 |
283 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA360392321 rs1246258453 |
284 | E>K | No |
ClinGen TOPMed |
|
|
rs781391894 CA3336363 |
286 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs144584603 CA3336361 |
287 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3336359 rs764093920 |
288 | N>Y | No |
ClinGen ExAC gnomAD |
|
|
rs201354427 COSM1071036 CA3336357 |
289 | V>I | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA3336356 rs140747495 |
291 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA360390960 rs1314154367 |
292 | K>N | No |
ClinGen gnomAD |
|
|
CA360390964 rs1356732396 |
292 | K>R | No |
ClinGen gnomAD |
|
|
rs374699547 CA3336326 |
293 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3336325 rs760022461 |
294 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs371730837 CA3336323 |
295 | K>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1343628362 CA360390885 |
296 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA3336320 rs777942986 |
300 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA360390823 rs1451863315 |
300 | D>N | No |
ClinGen gnomAD |
|
|
CA360390806 rs1186565268 |
301 | D>N | No |
ClinGen gnomAD |
|
|
rs778990691 CA3336316 |
303 | V>A | No |
ClinGen ExAC gnomAD |
|
|
COSM1071035 rs199734031 CA3336317 |
303 | V>I | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA360390736 rs1228332596 |
304 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1561341559 CA360390686 |
306 | K>N | No |
ClinGen Ensembl |
|
|
CA360390661 rs1298299916 |
308 | K>R | No |
ClinGen gnomAD |
|
|
CA3336312 rs781156139 |
309 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA3336314 rs751218019 |
309 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1213935250 CA360390650 |
309 | H>Y | No |
ClinGen gnomAD |
|
|
rs1345175979 CA360390637 |
310 | E>D | No |
ClinGen gnomAD |
|
|
rs1437383441 CA360390646 |
310 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1005653793 CA121826585 |
314 | W>R | No |
ClinGen gnomAD |
|
|
CA360390342 rs537556907 |
316 | D>H | No |
ClinGen 1000Genomes TOPMed |
|
|
rs537556907 CA121826565 |
316 | D>Y | No |
ClinGen 1000Genomes TOPMed |
|
|
rs749372407 CA3336275 |
318 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA360390328 rs148031256 |
318 | D>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3336276 rs148031256 |
318 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs770998749 CA3336273 |
322 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA121826524 rs1022139511 |
322 | S>P | No |
ClinGen TOPMed |
|
|
CA560884800 rs1460820474 |
322 | S>Y | No |
ClinGen gnomAD |
|
| TCGA novel | 323 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with P51946
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| CAK-ERCC2 complex | A protein complex formed by the association of the cyclin-dependent protein kinase activating kinase (CAK) holoenzyme complex with ERCC2. |
| cyclin-dependent protein kinase activating kinase holoenzyme complex | A cyclin-dependent kinase activating kinase complex capable of activating cyclin-dependent kinases by threonine phosphorylation, thus regulating cell cycle progression. consists of a kinase, cyclin and optional assembly factors, in human CDK7, CCNH and MNAT1. CAK activity is itself regulated throughout the cell cycle by T-loop phosphorylation of its kinase component (CDK7 in human). Phosphorylation of serine residues during mitosis inactivates the enzyme. Also capable of CAK phosphorylating the carboxyl-terminal domain (CTD) of RNA polymerase II and other transcription activating proteins, as part of the general transcription factor TFIIH. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| transcription factor TFIIH core complex | The 7 subunit core of TFIIH that is a part of either the general transcription factor holo-TFIIH or the nucleotide-excision repair factor 3 complex. In S. cerevisiae/humans the complex is composed of: Ssl2/XPB, Tfb1/p62, Tfb2/p52, Ssl1/p44, Tfb4/p34, Tfb5/p8 and Rad3/XPD. |
| transcription factor TFIIH holo complex | A complex that is capable of kinase activity directed towards the C-terminal Domain (CTD) of the largest subunit of RNA polymerase II and is essential for initiation at RNA polymerase II promoters in vitro. It is composed of the core TFIIH complex and the TFIIK complex. |
| transcription factor TFIIK complex | A transcription factor complex that forms part of the holo TFIIH complex. In Saccharomyces/human, TFIIK contains Ccl1p/Cyclin H, Tfb3p/MAT1 and Kin28p/CDK7. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| cyclin-dependent protein serine/threonine kinase regulator activity | Modulates the activity of a cyclin-dependent protein serine/threonine kinase, enzymes of the protein kinase family that are regulated through association with cyclins and other proteins. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| phosphorylation of RNA polymerase II C-terminal domain | The process of introducing a phosphate group on to an amino acid residue in the C-terminal domain of RNA polymerase II. Typically, this occurs during the transcription cycle and results in production of an RNA polymerase II enzyme where the carboxy-terminal domain (CTD) of the largest subunit is extensively phosphorylated, often referred to as hyperphosphorylated or the II(0) form. Specific types of phosphorylation within the CTD are usually associated with specific regions of genes, though there are exceptions. The phosphorylation state regulates the association of specific complexes such as the capping enzyme or 3'-RNA processing machinery to the elongating RNA polymerase complex. |
| protein stabilization | Any process involved in maintaining the structure and integrity of a protein and preventing it from degradation or aggregation. |
| regulation of cyclin-dependent protein serine/threonine kinase activity | Any process that modulates the frequency, rate or extent of cyclin-dependent protein serine/threonine kinase activity. |
| regulation of G1/S transition of mitotic cell cycle | Any signalling pathway that modulates the activity of a cell cycle cyclin-dependent protein kinase to modulate the switch from G1 phase to S phase of the mitotic cell cycle. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| transcription by RNA polymerase II | The synthesis of RNA from a DNA template by RNA polymerase II (RNAP II), originating at an RNA polymerase II promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs). |
| transcription initiation at RNA polymerase II promoter | A transcription initiation process that takes place at a RNA polymerase II gene promoter. Messenger RNAs (mRNA) genes, as well as some non-coding RNAs, are transcribed by RNA polymerase II. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3ZBL9 | CCNH | Cyclin-H | Bos taurus (Bovine) | PR |
| P24863 | CCNC | Cyclin-C | Homo sapiens (Human) | PR |
| O60563 | CCNT1 | Cyclin-T1 | Homo sapiens (Human) | EV |
| O75909 | CCNK | Cyclin-K | Homo sapiens (Human) | PR |
| Q8N1B3 | CCNQ | Cyclin-Q | Homo sapiens (Human) | PR |
| Q61458 | Ccnh | Cyclin-H | Mus musculus (Mouse) | PR |
| Q9R1A0 | Ccnh | Cyclin-H | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MYHNSSQKRH | WTFSSEEQLA | RLRADANRKF | RCKAVANGKV | LPNDPVFLEP | HEEMTLCKYY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EKRLLEFCSV | FKPAMPRSVV | GTACMYFKRF | YLNNSVMEYH | PRIIMLTCAF | LACKVDEFNV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SSPQFVGNLR | ESPLGQEKAL | EQILEYELLL | IQQLNFHLIV | HNPYRPFEGF | LIDLKTRYPI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LENPEILRKT | ADDFLNRIAL | TDAYLLYTPS | QIALTAILSS | ASRAGITMES | YLSESLMLKE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NRTCLSQLLD | IMKSMRNLVK | KYEPPRSEEV | AVLKQKLERC | HSAELALNVI | TKKRKGYEDD |
| 310 | 320 | ||||
| DYVSKKSKHE | EEEWTDDDLV | ESL |