P51813
Gene name |
BMX |
Protein name |
Cytoplasmic tyrosine-protein kinase BMX |
Names |
EC 2.7.10.2 , Bone marrow tyrosine kinase gene in chromosome X protein , Epithelial and endothelial tyrosine kinase , ETK , NTK38 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:660 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
|
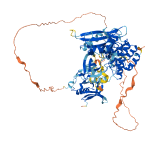
Descriptions
Autoinhibitory domains (AIDs)
Accessory elements
553-577 (Activation loop from InterPro)
Target domain |
417-675 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
Autoinhibited structure

Activated structure

458 variants for P51813
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs769401221 | 3 | T>I | No |
ExAC TOPMed |
|
| rs769401221 | 3 | T>K | No |
ExAC TOPMed |
|
| rs1295340360 | 4 | K>N | No | gnomAD | |
| rs367942269 | 5 | S>F | No |
ESP ExAC gnomAD |
|
| rs1044890948 | 5 | S>P | No | TOPMed | |
| rs1227560676 | 6 | I>T | No | gnomAD | |
| rs1264319329 | 8 | E>K | No | gnomAD | |
| rs371855296 | 9 | E>D | No |
ESP TOPMed |
|
| rs1923820921 | 10 | L>F | No | TOPMed | |
| rs773743068 | 12 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 14 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 14 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770720731 | 16 | Q>E | No |
ExAC gnomAD |
|
| rs1923822164 | 17 | Q>R | No | TOPMed | |
| COSM1315386 | 19 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774098459 | 23 | P>Q | No |
ExAC gnomAD |
|
| rs374784538 | 26 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1490545876 | 27 | K>E | No | TOPMed | |
| rs765370721 | 29 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs751155900 | 29 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1203354355 | 30 | L>F | No |
TOPMed gnomAD |
|
| rs1388114636 | 30 | L>R | No |
TOPMed gnomAD |
|
| rs762967277 | 38 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1234517546 | 42 | E>D | No | gnomAD | |
| rs751696072 | 43 | Y>F | No | ExAC | |
| rs1923826593 | 44 | D>E | No | Ensembl | |
| rs1416108873 | 47 | K>Q | No | gnomAD | |
|
rs763057245 COSM290142 |
48 | R>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs763057245 | 48 | R>M | No |
ExAC gnomAD |
|
| rs1312913843 | 48 | R>W | No | gnomAD | |
| rs775055084 | 50 | S>N | No |
1000Genomes ExAC gnomAD |
|
| COSM1118411 | 52 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1284788838 | 54 | S>F | No |
TOPMed gnomAD |
|
| rs1923859818 | 55 | I>T | No |
TOPMed gnomAD |
|
| rs375412828 | 55 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs767710151 | 58 | K>R | No |
ExAC gnomAD |
|
| rs1602324611 | 61 | R>K | No | Ensembl | |
| rs753014981 | 62 | C>W | No |
ExAC gnomAD |
|
| rs756497600 | 64 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| COSM1118412 | 68 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778455721 | 69 | E>K | No |
ExAC gnomAD |
|
|
rs774703027 COSM181596 |
72 | T>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1179167840 | 73 | P>L | No | gnomAD | |
| rs778275008 | 74 | V>A | No |
ExAC gnomAD |
|
| rs1433581281 | 75 | E>A | No | TOPMed | |
| rs1419463802 | 75 | E>K | No | gnomAD | |
| rs1923862732 | 77 | Q>R | No | TOPMed | |
| TCGA novel | 78 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs987183521 | 79 | P>S | No | Ensembl | |
| rs745602815 | 81 | Q>E | No |
ExAC gnomAD |
|
| rs1389419278 | 82 | I>V | No | gnomAD | |
| rs879056556 | 83 | V>A | No | gnomAD | |
| rs757699086 | 83 | V>I | No |
ExAC gnomAD |
|
| rs1923954261 | 84 | Y>C | No | Ensembl | |
| rs1405382906 | 85 | K>E | No | TOPMed | |
| TCGA novel | 87 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 87 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1171854947 | 88 | L>I | No | gnomAD | |
| rs1923955043 | 91 | V>A | No | Ensembl | |
| rs1457081076 | 92 | Y>C | No | gnomAD | |
| rs369144737 | 95 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs369144737 | 95 | N>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1923956150 | 97 | E>D | No | TOPMed | |
| rs914617893 | 97 | E>K | No | gnomAD | |
| rs779395852 | 98 | S>R | No |
ExAC gnomAD |
|
| rs150627254 | 99 | R>* | No |
ESP TOPMed |
|
|
COSM1184800 rs947474924 |
99 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs1923956911 | 100 | S>G | No | Ensembl | |
| TCGA novel | 105 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746549419 | 107 | Q>R | No |
ExAC gnomAD |
|
| rs1923957492 | 108 | K>N | No | TOPMed | |
| rs765493116 | 110 | I>V | No |
ExAC gnomAD |
|
| TCGA novel | 112 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757943077 | 112 | G>S | No |
ExAC gnomAD |
|
| rs1335673979 | 113 | N>K | No |
TOPMed gnomAD |
|
| rs372642389 | 115 | H>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs372642389 | 115 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs377713371 | 118 | V>L | No |
ESP TOPMed gnomAD |
|
| rs1445273890 | 122 | S>N | No | gnomAD | |
| rs1268607426 | 122 | S>R | No | Ensembl | |
| rs1924145439 | 123 | G>V | No | TOPMed | |
| rs1258473046 | 126 | V>G | No | Ensembl | |
| rs752611004 | 126 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs149800709 | 128 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
|
rs149800709 COSM422618 |
128 | G>W | urinary_tract [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| TCGA novel | 129 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1419609569 | 130 | F>Y | No | gnomAD | |
| rs749131478 | 131 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1158953515 | 133 | C>F | No | gnomAD | |
| rs1392131728 | 137 | C>F | No | TOPMed | |
| rs1924148469 | 140 | A>V | No | TOPMed | |
| rs2147108820 | 142 | G>E | No | Ensembl | |
| rs1319561771 | 144 | T>N | No | gnomAD | |
| rs765985964 | 145 | L>F | No |
1000Genomes ExAC gnomAD |
|
| COSM3560028 | 146 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200226983 | 149 | Y>F | No |
ExAC gnomAD |
|
| rs1409219021 | 153 | H>Q | No |
TOPMed gnomAD |
|
| rs1924237458 | 153 | H>Y | No | TOPMed | |
| rs1924238037 | 155 | A>S | No | Ensembl | |
| rs1924238037 | 155 | A>T | No | Ensembl | |
| rs753509328 | 155 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1450025000 | 158 | E>K | No |
TOPMed gnomAD |
|
| rs1024865323 | 160 | K>N | No | Ensembl | |
| COSM3844176 | 162 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1279885779 | 165 | T>I | No | gnomAD | |
| TCGA novel | 165 | T>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1924240587 | 166 | F>L | No | Ensembl | |
| rs1924240353 | 166 | F>L | No | Ensembl | |
| rs778907612 | 169 | R>G | No |
ExAC gnomAD |
|
| rs745615904 | 169 | R>K | No | Ensembl | |
| rs747210573 | 170 | V>A | No | ExAC | |
| rs747210573 | 170 | V>G | No | ExAC | |
| rs1205518576 | 172 | K>R | No | TOPMed | |
| rs1311621702 | 173 | I>T | No |
TOPMed gnomAD |
|
| rs1924500879 | 174 | P>L | No | Ensembl | |
| COSM1118414 | 174 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1305549193 | 175 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1924501372 | 175 | R>W | No | TOPMed | |
| TCGA novel | 176 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755198231 | 177 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1924501900 | 177 | V>F | No | TOPMed | |
| rs1924501900 | 177 | V>L | No | TOPMed | |
| rs1924502550 | 181 | K>E | No | Ensembl | |
| rs781573176 | 181 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs267606406 | 182 | M>I | No | Ensembl | |
| rs565126348 | 183 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1118415 | 184 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777767005 | 186 | S>F | No |
ExAC gnomAD |
|
| rs756208089 | 186 | S>T | No |
ExAC gnomAD |
|
| rs202245028 | 187 | S>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1924504600 | 188 | S>R | No | Ensembl | |
| rs1195837869 | 189 | T>A | No |
TOPMed gnomAD |
|
| rs1569222806 | 192 | A>G | No | Ensembl | |
| COSM1497217 | 193 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1449357667 | 194 | Y>H | No | gnomAD | |
| TCGA novel | 196 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746245000 | 197 | E>K | No |
ExAC gnomAD |
|
| rs1924506588 | 198 | S>L | No | TOPMed | |
| rs1924507068 | 200 | K>T | No | Ensembl | |
| rs1162894603 | 201 | N>D | No |
TOPMed gnomAD |
|
| COSM1118416 | 201 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771645389 | 203 | G>S | No |
ExAC gnomAD |
|
| rs976979984 | 204 | S>F | No |
TOPMed gnomAD |
|
| rs1327634458 | 204 | S>P | No |
TOPMed gnomAD |
|
| COSM488196 | 205 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1038666567 | 207 | P>L | No | TOPMed | |
| TCGA novel | 209 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1924509487 | 210 | S>G | No | TOPMed | |
| rs147917241 | 212 | S>I | No |
ExAC TOPMed gnomAD |
|
|
COSM73847 rs760301583 |
212 | S>R | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs147917241 | 212 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1924510233 | 213 | L>P | No | TOPMed | |
| rs139052738 | 214 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs199940605 | 214 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs139052738 RCV000903359 |
214 | A>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1274171514 | 216 | Y>C | No | gnomAD | |
| rs1203889554 | 218 | S>C | No | gnomAD | |
| rs1924512581 | 221 | K>E | No | Ensembl | |
| rs766149401 | 221 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1040042873 | 224 | Y>F | No |
TOPMed gnomAD |
|
| rs1324416054 | 225 | G>A | No | TOPMed | |
| COSM4108308 | 225 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752945588 | 226 | S>Y | No |
ExAC gnomAD |
|
| rs892165015 | 227 | Q>K | No | TOPMed | |
| rs1238266913 | 228 | P>L | No |
TOPMed gnomAD |
|
| rs1418301573 | 232 | M>I | No | gnomAD | |
| rs369745985 | 232 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs369745985 | 232 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1452700357 | 233 | Q>L | No | gnomAD | |
| rs1452700357 | 233 | Q>P | No | gnomAD | |
| COSM756713 | 236 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3560031 | 237 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1386993533 | 239 | D>G | No | gnomAD | |
| rs371837504 | 240 | F>L | No |
ESP ExAC gnomAD |
|
| rs371837504 | 240 | F>V | No |
ESP ExAC gnomAD |
|
| rs1569222981 | 242 | D>N | No | Ensembl | |
| rs779397369 | 243 | W>* | No |
ExAC gnomAD |
|
| rs1474913393 | 244 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1234732079 | 245 | Q>* | No |
TOPMed gnomAD |
|
| rs1234732079 | 245 | Q>E | No |
TOPMed gnomAD |
|
| rs199628600 | 245 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs199628600 | 245 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1003202379 | 246 | V>E | No | gnomAD | |
| rs1003202379 | 246 | V>G | No | gnomAD | |
| rs1316698478 | 246 | V>I | No | gnomAD | |
| rs1924519064 | 251 | S>I | No | Ensembl | |
| rs1924519064 | 251 | S>N | No | Ensembl | |
| rs1309563062 | 251 | S>R | No | gnomAD | |
| COSM4108309 | 253 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764236706 | 254 | S>G | No |
ExAC gnomAD |
|
| rs1924660077 | 254 | S>N | No | gnomAD | |
| rs1924660335 | 255 | S>N | No | TOPMed | |
| TCGA novel | 256 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1118418 | 257 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753849645 | 258 | V>A | No |
ExAC gnomAD |
|
| rs749267710 | 261 | S>C | No |
1000Genomes ExAC gnomAD |
|
| rs749267710 | 261 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs750905064 | 261 | S>N | No |
ExAC gnomAD |
|
| rs1924661888 | 264 | K>E | No |
TOPMed gnomAD |
|
| rs1252385673 | 267 | N>H | No |
TOPMed gnomAD |
|
| rs1223451888 | 268 | V>A | No | TOPMed | |
| rs2147118123 | 269 | N>K | No | Ensembl | |
| rs889888710 | 269 | N>T | No | TOPMed | |
| COSM1118419 | 274 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1118420 | 275 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1924663605 | 275 | I>N | No | TOPMed | |
| COSM3560032 | 277 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1393128794 | 277 | W>R | No | gnomAD | |
| rs758888055 | 280 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1189907523 | 283 | S>R | No | TOPMed | |
|
VAR_041674 rs35353387 |
284 | S>L | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1924708761 | 285 | S>T | No | TOPMed | |
| rs751881194 | 285 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM3844181 | 286 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1426155340 | 288 | E>K | No | gnomAD | |
| rs755287845 | 291 | L>P | No |
ExAC gnomAD |
|
| rs1400514787 | 292 | D>A | No | gnomAD | |
| rs1924710205 | 294 | Y>F | No | TOPMed | |
| rs1190640247 | 295 | D>E | No | gnomAD | |
| rs1924985310 | 298 | A>G | No | TOPMed | |
| rs1243441744 | 300 | N>I | No | gnomAD | |
|
COSM1118421 rs1924985813 |
301 | I>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs761589929 | 302 | S>A | No |
1000Genomes ExAC gnomAD |
|
| rs1924987138 | 308 | Q>E | No |
TOPMed gnomAD |
|
| rs1441960680 | 309 | L>F | No | TOPMed | |
| TCGA novel | 312 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM69835 | 313 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751992510 | 314 | G>E | No |
ExAC gnomAD |
|
| rs1925061440 | 315 | K>N | No | Ensembl | |
| rs755553413 | 315 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1471275479 | 320 | M>T | No | gnomAD | |
| rs923698155 | 320 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
|
COSM1118422 rs767957207 |
324 | S>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 324 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1177781301 | 328 | G>E | No |
TOPMed gnomAD |
|
| rs1228626782 | 330 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1380860780 | 331 | T>K | No | gnomAD | |
| rs755772429 | 332 | V>E | No |
ExAC gnomAD |
|
| rs777574174 | 334 | L>I | No |
ExAC gnomAD |
|
| rs1925064500 | 334 | L>S | No | Ensembl | |
| rs1320554964 | 339 | V>M | No |
TOPMed gnomAD |
|
|
COSM3800512 rs1601651847 |
340 | N>K | urinary_tract Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated TOPMed gnomAD NCI-TCGA Cosmic |
| rs756542747 | 344 | G>E | No |
ExAC gnomAD |
|
| rs866122314 | 348 | H>Y | No | Ensembl | |
| TCGA novel | 349 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756973630 | 350 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs753409287 | 350 | H>Y | No |
ExAC gnomAD |
|
| rs1013910902 | 351 | V>L | No |
TOPMed gnomAD |
|
| rs1013910902 | 351 | V>M | No |
TOPMed gnomAD |
|
| rs1175565787 | 353 | T>I | No | gnomAD | |
| rs1403243823 | 354 | N>K | No |
TOPMed gnomAD |
|
| COSM4820222 | 356 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1328014887 | 359 | L>S | No | gnomAD | |
| TCGA novel | 360 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1352553436 | 360 | Y>H | No | gnomAD | |
| rs747011166 | 362 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs747011166 | 362 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1231486655 | 364 | N>D | No | gnomAD | |
| rs1276083449 | 366 | C>Y | No | gnomAD | |
| TCGA novel | 373 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM457159 | 374 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1925229910 | 375 | H>Y | No | Ensembl | |
| rs1925230145 | 381 | S>L | No | TOPMed | |
| rs749435194 | 382 | A>G | No |
ExAC gnomAD |
|
|
rs1467580394 COSM3560033 |
384 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1925343048 | 387 | R>Q | No | TOPMed | |
| rs767793685 | 387 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs887128750 | 389 | R>C | No |
TOPMed gnomAD |
|
| rs1446519223 | 389 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM6118419 | 390 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754941859 | 391 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs754941859 | 391 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs754941859 | 391 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1375039619 | 393 | S>P | No | gnomAD | |
| rs780805113 | 394 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1310375412 | 395 | K>M | No | TOPMed | |
| rs1171195833 | 396 | A>T | No |
TOPMed gnomAD |
|
| rs1459615971 | 397 | N>H | No | gnomAD | |
| rs1391314844 | 400 | P>S | No |
TOPMed gnomAD |
|
|
COSM1490734 rs1384281513 |
401 | D>N | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs1601654516 | 401 | D>V | No | Ensembl | |
| rs1925346596 | 402 | S>A | No | TOPMed | |
| rs1156927059 | 404 | S>C | No | TOPMed | |
| rs772258045 | 404 | S>P | No |
ExAC gnomAD |
|
| rs775751232 | 406 | G>E | No | ExAC | |
| rs761221539 | 407 | N>D | No |
ExAC gnomAD |
|
| rs1451449698 | 407 | N>T | No | TOPMed | |
| COSM3913693 | 408 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201403849 | 409 | I>N | No |
1000Genomes ExAC gnomAD |
|
| rs1353133706 | 410 | W>* | No | gnomAD | |
| rs777479129 | 412 | L>P | No | ExAC | |
| rs1242272921 | 413 | K>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 415 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1313851206 | 415 | E>K | No | gnomAD | |
|
rs1046844113 COSM1466981 |
418 | T>A | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs1314137742 | 421 | K>E | No | TOPMed | |
| rs1037111073 | 422 | E>* | No |
TOPMed gnomAD |
|
| rs144679519 | 422 | E>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs144679519 | 422 | E>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1027257262 | 423 | L>P | No |
TOPMed gnomAD |
|
| rs1925404390 | 423 | L>V | No | Ensembl | |
| COSM3560034 | 424 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1354521022 | 428 | F>V | No | gnomAD | |
| rs1303127995 | 430 | V>M | No |
TOPMed gnomAD |
|
| rs780230726 | 432 | Q>H | No |
ExAC gnomAD |
|
| rs747098469 | 434 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs768976466 | 435 | K>E | No |
ExAC TOPMed |
|
| COSM4525290 | 441 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1420865167 | 442 | V>A | No |
TOPMed gnomAD |
|
| rs1418220685 | 444 | V>I | No | gnomAD | |
| rs1009730700 | 447 | I>N | No | TOPMed | |
| rs1009730700 | 447 | I>T | No | TOPMed | |
| rs1925408774 | 448 | K>Q | No | TOPMed | |
| rs1236633479 | 449 | E>Q | No | gnomAD | |
| rs1007023293 | 450 | G>S | No |
TOPMed gnomAD |
|
| rs773603922 | 452 | M>T | No |
ExAC gnomAD |
|
| rs770170556 | 452 | M>V | No |
ExAC gnomAD |
|
|
COSM298594 rs754428384 |
457 | F>L | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1425725331 | 460 | E>V | No | TOPMed | |
| rs1925411178 | 461 | A>V | No | Ensembl | |
| rs1396233126 | 463 | T>S | No | gnomAD | |
| rs1389973115 | 464 | M>T | No | gnomAD | |
| rs1414778887 | 465 | M>T | No | TOPMed | |
| rs1440812318 | 467 | L>F | No | gnomAD | |
| rs759995462 | 468 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs542606045 | 470 | P>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372808714 | 474 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1118424 | 475 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4912886 | 475 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs892209108 | 478 | V>A | No | Ensembl | |
| rs1925713367 | 481 | K>T | No | Ensembl | |
| rs2147137288 | 482 | E>K | No | 1000Genomes | |
|
rs1601661677 COSM1557400 COSM6186470 |
486 | Y>H | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl |
| rs767025241 | 489 | T>I | No |
ExAC gnomAD |
|
| rs1925714813 | 492 | I>T | No | TOPMed | |
| rs1489188708 | 493 | S>I | No |
TOPMed gnomAD |
|
| rs774895852 | 494 | N>S | No |
ExAC gnomAD |
|
| rs1260665188 | 495 | G>D | No | gnomAD | |
| rs764139510 | 497 | L>M | No |
ExAC gnomAD |
|
| rs763292985 | 498 | L>M | No | 1000Genomes | |
| rs753620703 | 500 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs757112613 | 503 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs201534309 | 504 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1288345731 | 505 | G>A | No |
TOPMed gnomAD |
|
| COSM3560035 | 505 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751804939 | 505 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1312781008 | 506 | K>Q | No | TOPMed | |
| TCGA novel | 507 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755193536 | 507 | G>R | No |
ExAC gnomAD |
|
| TCGA novel | 508 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs953291130 | 510 | P>L | No |
TOPMed gnomAD |
|
| rs1925718844 | 510 | P>S | No | TOPMed | |
| rs1925718844 | 510 | P>T | No | TOPMed | |
| rs781289361 | 513 | L>V | No |
ExAC gnomAD |
|
| COSM1118425 | 516 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1323399158 | 519 | D>N | No |
TOPMed gnomAD |
|
| rs1323399158 | 519 | D>Y | No |
TOPMed gnomAD |
|
| rs756580061 | 520 | V>I | No | ExAC | |
| rs1221736172 | 524 | M>V | No | gnomAD | |
| rs375810047 | 525 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1486983136 | 527 | L>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1925721674 | 528 | E>D | No | TOPMed | |
| rs1925721907 | 530 | H>Q | No | TOPMed | |
| rs1925722132 | 531 | Q>R | No | TOPMed | |
|
COSM4108312 rs1925722605 |
534 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs749852426 | 534 | H>Y | No |
ExAC gnomAD |
|
| rs1180445272 | 535 | R>Q | No | gnomAD | |
| rs758831876 | 535 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1925771600 | 538 | A>V | No | TOPMed | |
| rs1601662996 | 539 | A>S | No | Ensembl | |
| rs759748843 | 540 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs374705811 | 540 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs1473294151 | 544 | V>A | No | gnomAD | |
| rs149547172 | 546 | R>G | No |
ESP TOPMed gnomAD |
|
| rs1349359949 | 546 | R>T | No |
TOPMed gnomAD |
|
| rs1441660683 | 547 | D>G | No | gnomAD | |
| rs1925773944 | 548 | L>I | No | TOPMed | |
| rs777880915 | 549 | C>W | No |
ExAC gnomAD |
|
| COSM6005180 | 550 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1412977050 | 550 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
TCGA novel rs754312789 |
555 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
| rs1925775387 | 557 | M>K | No | TOPMed | |
| rs1925775387 | 557 | M>R | No | TOPMed | |
| TCGA novel | 558 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1118427 | 562 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1330394833 | 563 | D>N | No |
TOPMed gnomAD |
|
| rs1925968894 | 563 | D>V | No | TOPMed | |
| rs760927809 | 565 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs760927809 | 565 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs764120775 | 567 | V>I | No |
ExAC gnomAD |
|
| rs1034065641 | 568 | S>R | No | Ensembl | |
| rs144206884 | 571 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs994609022 | 572 | T>R | No | TOPMed | |
| rs765665248 | 573 | K>N | No |
ExAC TOPMed gnomAD |
|
|
COSM1118428 rs751005076 |
575 | P>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs747676627 | 578 | W>* | No | Ensembl | |
| rs1925972062 | 579 | S>T | No | Ensembl | |
| rs1925972296 | 589 | Y>H | No | TOPMed | |
| COSM756711 | 593 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780513020 | 593 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs746626126 | 595 | V>I | No |
ExAC gnomAD |
|
| rs746626126 | 595 | V>L | No |
ExAC gnomAD |
|
| rs780798273 | 596 | W>R | No |
ExAC gnomAD |
|
| rs1926108706 | 600 | I>M | No | Ensembl | |
| rs2147144417 | 600 | I>N | No | Ensembl | |
| rs780990360 | 602 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs889949576 | 604 | E>K | No | TOPMed | |
| rs1191799164 | 607 | S>G | No | gnomAD | |
| rs1239574626 | 607 | S>I | No |
TOPMed gnomAD |
|
| rs1265550312 | 607 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs755560266 | 609 | G>E | No |
ExAC gnomAD |
|
| rs755560266 | 609 | G>V | No |
ExAC gnomAD |
|
| rs777387731 | 614 | D>E | No |
ExAC gnomAD |
|
| rs1364382238 | 614 | D>N | No | Ensembl | |
| COSM1466982 | 615 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1445530814 | 616 | Y>* | No | gnomAD | |
| rs1007742292 | 616 | Y>C | No | TOPMed | |
| rs749140949 | 618 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs749140949 | 618 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1164564596 | 619 | S>P | No | gnomAD | |
| rs773960824 | 621 | V>A | No |
ExAC gnomAD |
|
| rs1418508053 | 621 | V>L | No |
TOPMed gnomAD |
|
| rs143079475 | 622 | V>A | No |
ESP ExAC |
|
| rs568883721 | 630 | R>S | No | Ensembl | |
| rs762070386 | 633 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs776820082 | 633 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2147144516 | 634 | P>T | No | Ensembl | |
| rs765195659 | 635 | H>Q | No |
ExAC gnomAD |
|
| COSM3560036 | 635 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs756397275 COSM457160 |
638 | S>L | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| COSM1118429 | 638 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1029219370 | 641 | I>N | No | Ensembl | |
| rs752180539 | 645 | M>R | No |
ExAC gnomAD |
|
| rs752180539 | 645 | M>T | No |
ExAC gnomAD |
|
| rs768047252 | 650 | H>L | No |
ExAC TOPMed gnomAD |
|
| rs752370479 | 650 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs768047252 | 650 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1368942344 | 651 | E>D | No | gnomAD | |
| rs1189561826 | 651 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2147150353 | 655 | K>E | No | Ensembl | |
| rs770977325 | 655 | K>N | No |
ExAC gnomAD |
|
|
rs1926422844 COSM3035184 |
656 | R>C | liver Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1243725193 | 656 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 657 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1926423289 | 658 | T>A | No | Ensembl | |
| TCGA novel | 658 | T>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 658 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1451651014 | 661 | Q>* | No |
TOPMed gnomAD |
|
| rs1176309936 | 662 | L>F | No | gnomAD | |
| rs760164275 | 664 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs767957587 | 666 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1926424717 | 668 | P>S | No | TOPMed | |
| rs371758778 | 669 | L>P | No |
ESP TOPMed |
|
| rs1375536026 | 670 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
COSM21256 rs1029888196 VAR_041675 |
670 | R>W | lung Variant assessed as Somatic; MODERATE impact. urinary_tract a lung large cell carcinoma sample; somatic mutation [Cosmic, NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic cosmic curated UniProt NCI-TCGA TOPMed dbSNP gnomAD |
| rs1334381378 | 673 | D>N | No |
TOPMed gnomAD |
|
| TCGA novel | 673 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1173504581 | 675 | H>Q | No | gnomAD |
No associated diseases with P51813
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction |
| protein tyrosine kinase activity | Catalysis of the reaction |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigens produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| B cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a B cell. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mesoderm development | The process whose specific outcome is the progression of the mesoderm over time, from its formation to the mature structure. The mesoderm is the middle germ layer that develops into muscle, bone, cartilage, blood and connective tissue. |
| phosphatidylinositol biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidylinositol, any glycophospholipid in which the sn-glycerol 3-phosphate residue is esterified to the 1-hydroxyl group of 1D-myo-inositol. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| T cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a T cell. |
86 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0JNB0 | FYN | Tyrosine-protein kinase Fyn | Bos taurus (Bovine) | SS |
| Q0VBZ0 | CSK | Tyrosine-protein kinase CSK | Bos taurus (Bovine) | SS |
| Q3ZC95 | BTK | Tyrosine-protein kinase | Bos taurus (Bovine) | EV SS |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| P00523 | SRC | Proto-oncogene tyrosine-protein kinase Src | Gallus gallus (Chicken) | EV |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| Q75R65 | JAK2 | Tyrosine-protein kinase JAK2 | Gallus gallus (Chicken) | SS |
| Q24592 | hop | Tyrosine-protein kinase hopscotch | Drosophila melanogaster (Fruit fly) | PR |
| Q9V9J3 | Src42A | Tyrosine-protein kinase Src42A | Drosophila melanogaster (Fruit fly) | SS |
| P00528 | Src64B | Tyrosine-protein kinase Src64B | Drosophila melanogaster (Fruit fly) | SS |
| P08630 | Btk | Tyrosine-protein kinase Btk | Drosophila melanogaster (Fruit fly) | SS |
| P23458 | JAK1 | Tyrosine-protein kinase JAK1 | Homo sapiens (Human) | SS |
| O60674 | JAK2 | Tyrosine-protein kinase JAK2 | Homo sapiens (Human) | EV |
| P52333 | JAK3 | Tyrosine-protein kinase JAK3 | Homo sapiens (Human) | SS |
| P29597 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | Homo sapiens (Human) | EV |
| P07947 | YES1 | Tyrosine-protein kinase Yes | Homo sapiens (Human) | SS |
| P09769 | FGR | Tyrosine-protein kinase Fgr | Homo sapiens (Human) | SS |
| P41240 | CSK | Tyrosine-protein kinase CSK | Homo sapiens (Human) | SS |
| P42680 | TEC | Tyrosine-protein kinase Tec | Homo sapiens (Human) | SS |
| P51451 | BLK | Tyrosine-protein kinase Blk | Homo sapiens (Human) | SS |
| Q08881 | ITK | Tyrosine-protein kinase ITK/TSK | Homo sapiens (Human) | EV |
| P43405 | SYK | Tyrosine-protein kinase SYK | Homo sapiens (Human) | EV |
| P43403 | ZAP70 | Tyrosine-protein kinase ZAP-70 | Homo sapiens (Human) | EV |
| Q13882 | PTK6 | Protein-tyrosine kinase 6 | Homo sapiens (Human) | EV |
| P07948 | LYN | Tyrosine-protein kinase Lyn | Homo sapiens (Human) | SS |
| P06241 | FYN | Tyrosine-protein kinase Fyn | Homo sapiens (Human) | SS |
| P12931 | SRC | Proto-oncogene tyrosine-protein kinase Src | Homo sapiens (Human) | EV |
| P06239 | LCK | Tyrosine-protein kinase Lck | Homo sapiens (Human) | EV |
| P08631 | HCK | Tyrosine-protein kinase HCK | Homo sapiens (Human) | EV |
| P42685 | FRK | Tyrosine-protein kinase FRK | Homo sapiens (Human) | EV |
| Q06187 | BTK | Tyrosine-protein kinase BTK | Homo sapiens (Human) | EV |
| P42679 | MATK | Megakaryocyte-associated tyrosine-protein kinase | Homo sapiens (Human) | SS |
| Q14289 | PTK2B | Protein-tyrosine kinase 2-beta | Homo sapiens (Human) | PR |
| Q05397 | PTK2 | Focal adhesion kinase 1 | Homo sapiens (Human) | EV |
| Q13470 | TNK1 | Non-receptor tyrosine-protein kinase TNK1 | Homo sapiens (Human) | PR |
| Q07912 | TNK2 | Activated CDC42 kinase 1 | Homo sapiens (Human) | EV |
| P16591 | FER | Tyrosine-protein kinase Fer | Homo sapiens (Human) | PR |
| Q6J9G0 | STYK1 | Tyrosine-protein kinase STYK1 | Homo sapiens (Human) | PR |
| P42684 | ABL2 | Tyrosine-protein kinase ABL2 | Homo sapiens (Human) | SS |
| P00519 | ABL1 | Tyrosine-protein kinase ABL1 | Homo sapiens (Human) | EV |
| Q9R117 | Tyk2 | Non-receptor tyrosine-protein kinase TYK2 | Mus musculus (Mouse) | SS |
| P08103 | Hck | Tyrosine-protein kinase HCK | Mus musculus (Mouse) | SS |
| P16277 | Blk | Tyrosine-protein kinase Blk | Mus musculus (Mouse) | SS |
| Q62270 | Srms | Tyrosine-protein kinase Srms | Mus musculus (Mouse) | SS |
| Q64434 | Ptk6 | Protein-tyrosine kinase 6 | Mus musculus (Mouse) | SS |
| P05480 | Src | Proto-oncogene tyrosine-protein kinase Src | Mus musculus (Mouse) | EV |
| P14234 | Fgr | Tyrosine-protein kinase Fgr | Mus musculus (Mouse) | SS |
| P35991 | Btk | Tyrosine-protein kinase BTK | Mus musculus (Mouse) | EV |
| P41241 | Csk | Tyrosine-protein kinase CSK | Mus musculus (Mouse) | EV |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | Mus musculus (Mouse) | EV |
| Q62137 | Jak3 | Tyrosine-protein kinase JAK3 | Mus musculus (Mouse) | SS |
| Q62120 | Jak2 | Tyrosine-protein kinase JAK2 | Mus musculus (Mouse) | EV |
| P06240 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Mus musculus (Mouse) | SS |
| P24604 | Tec | Tyrosine-protein kinase Tec | Mus musculus (Mouse) | SS |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | Mus musculus (Mouse) | SS |
| P39688 | Fyn | Tyrosine-protein kinase Fyn | Mus musculus (Mouse) | SS |
| P52332 | Jak1 | Tyrosine-protein kinase JAK1 | Mus musculus (Mouse) | SS |
| Q03526 | Itk | Tyrosine-protein kinase ITK/TSK | Mus musculus (Mouse) | SS |
| P41242 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q922K9 | Frk | Tyrosine-protein kinase FRK | Mus musculus (Mouse) | SS |
| A1Y2K1 | FYN | Tyrosine-protein kinase Fyn | Sus scrofa (Pig) | SS |
| O19064 | JAK2 | Tyrosine-protein kinase JAK2 | Sus scrofa (Pig) | SS |
| Q62662 | Frk | Tyrosine-protein kinase FRK | Rattus norvegicus (Rat) | SS |
| Q62844 | Fyn | Tyrosine-protein kinase Fyn | Rattus norvegicus (Rat) | SS |
| Q07014 | Lyn | Tyrosine-protein kinase Lyn | Rattus norvegicus (Rat) | SS |
| P50545 | Hck | Tyrosine-protein kinase HCK | Rattus norvegicus (Rat) | SS |
| Q9WUD9 | Src | Proto-oncogene tyrosine-protein kinase Src | Rattus norvegicus (Rat) | SS |
| Q01621 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Rattus norvegicus (Rat) | SS |
| Q6P6U0 | Fgr | Tyrosine-protein kinase Fgr | Rattus norvegicus (Rat) | SS |
| Q62689 | Jak2 | Tyrosine-protein kinase JAK2 | Rattus norvegicus (Rat) | SS |
| Q63272 | Jak3 | Tyrosine-protein kinase JAK3 | Rattus norvegicus (Rat) | SS |
| P32577 | Csk | Tyrosine-protein kinase CSK | Rattus norvegicus (Rat) | SS |
| P41243 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Rattus norvegicus (Rat) | SS |
| F1LM93 | Yes1 | Tyrosine-protein kinase Yes | Rattus norvegicus (Rat) | SS |
| O45539 | src-2 | Tyrosine protein-kinase src-2 | Caenorhabditis elegans | SS |
| G5ECJ6 | csk-1 | Tyrosine-protein kinase csk-1 | Caenorhabditis elegans | SS |
| G5EE56 | src-1 | Tyrosine protein-kinase src-1 | Caenorhabditis elegans | SS |
| A1A5H8 | yes1 | Tyrosine-protein kinase yes | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F1RDG9 | fynb | Tyrosine-protein kinase fynb | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O12990 | jak1 | Tyrosine-protein kinase JAK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q1JPZ3 | src | Proto-oncogene tyrosine-protein kinase Src | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6EWH2 | fyna | Tyrosine-protein kinase fyna | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDTKSILEEL | LLKRSQQKKK | MSPNNYKERL | FVLTKTNLSY | YEYDKMKRGS | RKGSIEIKKI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RCVEKVNLEE | QTPVERQYPF | QIVYKDGLLY | VYASNEESRS | QWLKALQKEI | RGNPHLLVKY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| HSGFFVDGKF | LCCQQSCKAA | PGCTLWEAYA | NLHTAVNEEK | HRVPTFPDRV | LKIPRAVPVL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KMDAPSSSTT | LAQYDNESKK | NYGSQPPSSS | TSLAQYDSNS | KKIYGSQPNF | NMQYIPREDF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PDWWQVRKLK | SSSSSEDVAS | SNQKERNVNH | TTSKISWEFP | ESSSSEEEEN | LDDYDWFAGN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ISRSQSEQLL | RQKGKEGAFM | VRNSSQVGMY | TVSLFSKAVN | DKKGTVKHYH | VHTNAENKLY |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LAENYCFDSI | PKLIHYHQHN | SAGMITRLRH | PVSTKANKVP | DSVSLGNGIW | ELKREEITLL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KELGSGQFGV | VQLGKWKGQY | DVAVKMIKEG | SMSEDEFFQE | AQTMMKLSHP | KLVKFYGVCS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KEYPIYIVTE | YISNGCLLNY | LRSHGKGLEP | SQLLEMCYDV | CEGMAFLESH | QFIHRDLAAR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| NCLVDRDLCV | KVSDFGMTRY | VLDDQYVSSV | GTKFPVKWSA | PEVFHYFKYS | SKSDVWAFGI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LMWEVFSLGK | QPYDLYDNSQ | VVLKVSQGHR | LYRPHLASDT | IYQIMYSCWH | ELPEKRPTFQ |
| 670 | |||||
| QLLSSIEPLR | EKDKH |