P51812
Gene name |
RPS6KA3 (ISPK1, MAPKAPK1B, RSK2) |
Protein name |
Ribosomal protein S6 kinase alpha-3 |
Names |
S6K-alpha-3, 90 kDa ribosomal protein S6 kinase 3, p90-RSK 3, p90RSK3, Insulin-stimulated protein kinase 1, ISPK-1, MAP kinase-activated protein kinase 1b, MAPK-activated protein kinase 1b, MAPKAP kinase 1b, MAPKAPK-1b, Ribosomal S6 kinase 2, RSK-2, pp90RSK2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6197 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
RIBOSOMAL PROTEIN S6 KINASE (PTHR24351) |
Descriptions
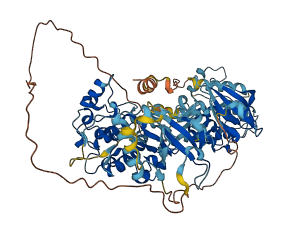
The 90-kDa ribosomal S6 kinase 2 (RSK2) is broadly expressed in response to growth factors, peptide hormones, neurotransmitters, chemokines and other stimuli. RSK2 is a serine/threonine kinase containing two distinct catalytically functional kinase domains connected by a linker region. The C-terminal domain (CTD) phosphorylates the linker region and regulates the N-terminal domain, which phosphorylates various substrates. The scaffold of the RSK2 CTD is stabilized by the αL-helix, which is located underneath the catalytic cleft and embedded in the kinase scaffold. On the basis of this structure of the CTD of RSK2, ERKs are likely to be involved in abolishing the autoinhibitory function of the CTD. The ERKs binding site (residues 726-735) is located at the RSK2 C terminus close to the αL-helix (residues 697-712). Efficient activation of RSK2 requires interaction with extracellular signal-regulated protein kinases (ERKs) at a docking site in the RSK2 C terminus (residues 726-735) and subsequent phosphorylation of Thr-577 in the CTD T-activation loop.
Autoinhibitory domains (AIDs)
Target domain |
422-679 (Protein kinase domain) |
Relief mechanism |
PTM, Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
210-233 (Activation loop from InterPro)
Target domain |
72-388 (N-terminal catalytic domain of the Serine/Threonine Kinase, 90 kDa ribosomal protein S6 kinase) |
Relief mechanism |
|
Assay |
|
560-583 (Activation loop from InterPro)
Target domain |
402-740 (C-terminal catalytic domain of the Serine/Threonine Kinase, Ribosomal S6 kinase 2, also called 90kDa ribosomal protein S6 kinase 3 or Ribosomal protein S6 kinase alpha-3) |
Relief mechanism |
|
Assay |
|
References
- Li D et al. (2012) "Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1", Acta crystallographica. Section D, Biological crystallography, 68, 680-5
- Li D et al. (2013) "The prometastatic ribosomal S6 kinase 2-cAMP response element-binding protein (RSK2-CREB) signaling pathway up-regulates the actin-binding protein fascin-1 to promote tumor metastasis", The Journal of biological chemistry, 288, 32528-32538
- Malakhova M et al. (2008) "Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2", Nature structural & molecular biology, 15, 112-3
- Poteet-Smith CE et al. (1999) "Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family", The Journal of biological chemistry, 274, 22135-8
Autoinhibited structure

Activated structure

13 structures for P51812
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4D9T | X-ray | 240 A | A | 399-740 | PDB |
| 4D9U | X-ray | 240 A | A | 399-740 | PDB |
| 4JG6 | X-ray | 260 A | A | 399-740 | PDB |
| 4JG7 | X-ray | 300 A | A | 399-740 | PDB |
| 4JG8 | X-ray | 310 A | A | 399-740 | PDB |
| 4NUS | X-ray | 239 A | A | 39-359 | PDB |
| 4NW5 | X-ray | 194 A | A | 39-359 | PDB |
| 4NW6 | X-ray | 174 A | A | 39-359 | PDB |
| 5D9K | X-ray | 255 A | A/B | 39-366 | PDB |
| 5D9L | X-ray | 215 A | A | 39-359 | PDB |
| 7OPO | X-ray | 275 A | A/C/E/G/I/K | 39-351 | PDB |
| 8EQ5 | X-ray | 180 A | A | 46-346 | PDB |
| AF-P51812-F1 | Predicted | AlphaFoldDB |
199 variants for P51812
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001251296 rs2068650679 RCV001819956 |
71 | L>* | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
VAR_006189 CA255955 RCV000012417 rs122454124 |
75 | G>V | Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome [Ensembl, UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA255957 rs122454126 VAR_006190 RCV000012419 |
82 | V>F | Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome [Ensembl, UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt TOPMed dbSNP |
|
RCV000703246 CA412511525 rs1569232184 |
99 | M>V | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000578332 rs1555943492 |
110 | R>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001257663 COSM1119115 RCV000660244 rs1555943484 CA412511086 RCV001796175 |
110 | R>* | Intellectual disability endometrium Coffin-Lowry syndrome breast [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
COSM1625780 rs1555943479 CA412511061 RCV000660245 |
112 | R>* | liver Coffin-Lowry syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
CA255959 rs122454127 RCV000012421 VAR_006191 COSM295761 |
114 | R>W | Coffin-lowry syndrome (cls) large_intestine CLS Coffin-Lowry syndrome breast [Ensembl, Cosmic, UniProt, ClinVar] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl dbSNP |
|
rs387906703 VAR_065892 RCV000022823 CA128777 |
115 | T>S | Intellectual disability, X-linked 19 Mental retardation, x-linked 19 (mrx19) XLID19 [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000012422 rs1603426295 |
151 | R>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs398122813 RCV000022822 VAR_065893 |
152 | G>missing | Intellectual disability, X-linked 19 XLID19 [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP |
|
CA16043585 rs1057518914 RCV000415122 |
178 | A>G | Hypertelorism [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000012426 VAR_065894 CA255963 rs122454130 |
189 | I>K | Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome [Ensembl, UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1325953089 CA412508767 RCV000677731 |
210 | T>I | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000625970 CA412508620 rs1555939456 COSM1625779 |
216 | K>E | liver Coffin-Lowry syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
VAR_006194 rs879027948 CA327475781 |
225 | A>V | CLS [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
VAR_006195 CA255956 rs122454125 RCV000012418 |
227 | S>A | Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome [Ensembl, UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1569216043 RCV000707723 CA412508272 |
236 | A>V | Coffin-lowry syndrome (cls) Coffin-Lowry syndrome [Ensembl, ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000622430 rs1555939377 CA412508251 |
238 | E>* | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1603425338 RCV000990500 CA412508072 COSM250743 |
250 | D>N | liver Coffin-Lowry syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
rs1569215936 RCV000690930 CA412508024 |
252 | W>R | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000660246 CA412507978 rs1555939335 |
255 | G>D | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs122454131 VAR_065896 CA255964 RCV000012432 |
268 | F>S | Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome [Ensembl, UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001266850 rs2067717120 |
297 | S>N | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000660248 rs1555933769 |
300 | R>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1603422403 RCV000789047 CA412518228 |
311 | L>* | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001253583 rs2067702700 |
320 | E>* | Intellectual disability, X-linked 19 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2067699201 RCV001257664 |
337 | R>G | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
CA121631 RCV000012425 rs122454129 VAR_065897 |
383 | R>W | Intellectual disability, X-linked 19 Mental retardation, x-linked 19 (mrx19) XLID19; kinase activity is decreased but not abolished [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000764872 rs765914103 CA10366139 RCV000591676 |
413 | R>G | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
RCV000193195 rs797045920 |
435 | V>missing | Intellectual disability, X-linked 19 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000660249 rs1160828151 CA412516189 |
436 | C>* | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000012433 rs1603420690 |
477 | I>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2067475470 RCV001252610 |
503 | D>H | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000626867 rs1555928716 CA412514831 |
508 | Q>* | Global developmental delay [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001267209 rs2067399344 |
538 | R>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000624534 rs1555927575 CA412514291 |
545 | I>V | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000624151 CA412514136 rs1555927554 |
567 | Q>* | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA412513996 rs1555927532 RCV000505211 |
588 | E>Q | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001250664 rs2067342745 |
608 | L>R | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000686752 rs1569190602 |
614 | G>* | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412513608 rs1555926370 RCV000660637 |
626 | P>L | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1085307639 RCV001249648 CA412513571 RCV000489984 |
632 | R>* | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1555924704 RCV000660250 |
655 | L>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2067184984 RCV001265989 |
657 | S>* | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs122454128 CA255960 RCV000012423 |
689 | Q>* | Coffin-lowry syndrome (cls) Coffin-Lowry syndrome [Ensembl, ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000012429 rs1603417213 |
715 | S>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA207224 RCV000193624 RCV000595652 rs144984628 RCV000721043 |
723 | R>H | Intellectual disability, X-linked 19 History of neurodevelopmental disorder [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs28935171 RCV000413967 RCV000012424 RCV001266366 RCV001257622 CA255962 VAR_006197 |
729 | R>Q | Intellectual disability Coffin-lowry syndrome (cls) CLS Coffin-Lowry syndrome Inborn genetic diseases [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000714792 rs1555924331 CA412511813 RCV000660251 |
729 | R>W | Intellectual disability, X-linked 19 Coffin-Lowry syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000790412 rs1603417191 |
735 | I>missing | Coffin-Lowry syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412510906 rs1227902305 |
2 | P>L | No |
ClinGen gnomAD |
|
|
CA412510902 rs1362995147 |
3 | L>V | No |
ClinGen gnomAD |
|
|
CA412510820 rs1429116502 |
6 | L>P | No |
ClinGen gnomAD |
|
|
rs1365070717 CA412510799 |
7 | A>G | No |
ClinGen gnomAD |
|
|
rs1210808520 CA412510710 |
10 | W>* | No |
ClinGen Ensembl |
|
|
rs982750268 CA327481713 |
14 | A>V | No |
ClinGen TOPMed |
|
|
CA412510513 rs1327104134 |
16 | E>D | No |
ClinGen gnomAD |
|
|
CA412510475 rs1463499917 |
17 | S>N | No |
ClinGen gnomAD |
|
|
CA412510448 rs1391562214 |
18 | P>L | No |
ClinGen gnomAD |
|
|
CA412510451 rs1391562214 |
18 | P>R | No |
ClinGen gnomAD |
|
|
rs1417378508 CA412510457 |
18 | P>S | No |
ClinGen gnomAD |
|
|
rs867635523 CA327481711 |
20 | D>N | No |
ClinGen Ensembl |
|
|
rs996264009 CA327481710 COSM251408 |
21 | S>I | liver [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs996264009 CA412510392 |
21 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
CA412510379 rs1421289336 |
22 | A>S | No |
ClinGen gnomAD |
|
|
CA412510357 rs1391619408 |
23 | E>D | No |
ClinGen TOPMed |
|
|
CA10366346 rs756013694 |
28 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10366345 rs752749317 |
32 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs767431175 CA10366344 |
33 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA412515439 rs1487389355 |
33 | M>V | No |
ClinGen gnomAD |
|
|
rs56218010 CA148271 VAR_006188 |
38 | I>S | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA10366343 rs751040058 |
41 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA16043738 RCV000415809 rs1057519103 |
41 | Q>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412513309 rs1187541060 |
46 | S>N | No |
ClinGen gnomAD |
|
|
RCV000118191 CA231462 rs140987045 |
47 | I>V | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA412513248 rs1319564910 |
50 | I>M | No |
ClinGen TOPMed |
|
|
rs777093366 CA10366318 |
51 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777093366 CA10366319 |
51 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs398124175 RCV000081154 |
62 | K>missing | No |
ClinVar dbSNP |
|
|
CA412513012 rs1334873346 |
65 | P>L | No |
ClinGen gnomAD |
|
|
RCV000425960 CA16609164 rs1057520540 |
68 | F>L | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs886041331 CA10603428 RCV000375901 |
69 | E>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1274506228 CA412512939 |
70 | L>F | No |
ClinGen gnomAD |
|
|
rs1242083152 CA412512936 |
70 | L>R | No |
ClinGen gnomAD |
|
|
rs1064793356 CA16621297 RCV000486841 |
81 | K>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA327477002 rs122454126 |
82 | V>I | Coffin-lowry syndrome (cls) [Ensembl] | No |
ClinGen TOPMed |
|
rs1555948447 RCV000522959 CA412511744 |
87 | K>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs376080865 CA10366307 |
88 | I>M | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1339757713 CA412511596 |
94 | R>K | No |
ClinGen gnomAD |
|
|
rs373435639 CA10366289 |
113 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
CA16043245 RCV000414595 rs1057517947 |
119 | R>P | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
RCV001008403 rs1603426296 |
148 | D>missing | No |
ClinVar dbSNP |
|
|
rs1165081832 CA412509848 |
186 | S>N | No |
ClinGen gnomAD |
|
|
CA10366256 rs369498308 |
196 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV000330683 rs886041330 |
202 | D>missing | No |
ClinVar dbSNP |
|
|
RCV000598903 rs1555939921 |
204 | E>missing | No |
ClinVar dbSNP |
|
|
RCV000428435 CA16608808 rs1057523854 |
206 | H>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1325953089 CA412508771 |
210 | T>K | No |
ClinGen gnomAD |
|
|
CA10606024 rs886043847 RCV000319579 |
211 | D>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412508762 rs1347633410 |
211 | D>N | No |
ClinGen gnomAD |
|
|
rs1603425362 RCV001008777 |
215 | S>missing | No |
ClinVar dbSNP |
|
|
rs1428809534 CA412508569 |
219 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA412508552 rs1163126922 |
220 | D>N | No |
ClinGen gnomAD |
|
|
rs1603425359 CA412508527 |
221 | H>R | No |
ClinGen Ensembl |
|
|
CA327475782 rs1037326610 |
225 | A>T | No |
ClinGen Ensembl |
|
|
rs398124178 RCV000081157 |
240 | V>missing | No |
ClinVar dbSNP |
|
|
RCV000414370 rs1057518020 CA16043302 |
240 | V>I | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1431378578 CA412508181 |
242 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1431378578 CA412508178 |
242 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1603425345 CA412508173 |
243 | R>* | No |
ClinGen Ensembl |
|
|
rs201276916 CA327475780 |
246 | T>A | No |
ClinGen 1000Genomes TOPMed |
|
|
rs891163492 CA327475779 |
247 | Q>H | No |
ClinGen Ensembl |
|
|
RCV000478197 rs1064793597 |
272 | D>missing | No |
ClinVar dbSNP |
|
|
rs746443280 CA10366214 |
272 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA412518354 rs1314565638 |
293 | P>A | No |
ClinGen gnomAD |
|
|
rs1314565638 CA412518353 |
293 | P>S | No |
ClinGen gnomAD |
|
|
CA10366204 rs765189733 |
295 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1603422409 CA412518306 RCV001007968 |
300 | R>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA358896 rs869320705 |
305 | R>* | No |
ClinGen Ensembl |
|
|
RCV000658440 CA412518250 rs1555933752 |
308 | A>E | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412518193 rs1356590482 |
315 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1356590482 CA412518192 |
315 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA10366191 rs753024493 |
325 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA412518103 rs1242203087 |
327 | F>L | No |
ClinGen gnomAD |
|
|
rs758308122 CA10366189 |
329 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA327474916 rs868646575 |
339 | E>G | No |
ClinGen Ensembl |
|
|
CA412517948 rs1225839586 |
342 | P>L | No |
ClinGen TOPMed |
|
|
CA412517861 rs1418802319 |
348 | T>M | No |
ClinGen TOPMed gnomAD |
|
|
rs774729580 CA10366175 |
356 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA10366170 rs748206319 |
367 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA412516910 rs1273664992 |
389 | A>P | No |
ClinGen gnomAD |
|
|
rs1200950094 CA412516887 |
392 | S>L | No |
ClinGen gnomAD |
|
|
CA412516892 rs1157623710 |
392 | S>T | No |
ClinGen TOPMed |
|
|
rs1347117859 CA412516883 |
393 | D>G | No |
ClinGen gnomAD |
|
|
rs138802948 CA10366158 |
396 | S>N | No |
ClinGen ESP ExAC gnomAD |
|
|
rs766835360 CA10366157 |
397 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs761361669 CA327474835 |
399 | M>V | No |
ClinGen Ensembl |
|
|
CA412516814 rs1289181052 |
403 | G>S | No |
ClinGen gnomAD |
|
|
rs144961960 CA10366154 |
407 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA412516605 rs1242790695 |
414 | N>S | No |
ClinGen TOPMed |
|
|
CA412516593 rs1490252916 |
415 | S>G | No |
ClinGen TOPMed |
|
|
rs148050184 CA412516580 |
416 | I>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1449278063 CA412516568 |
416 | I>M | No |
ClinGen gnomAD |
|
|
rs148050184 CA10366137 VAR_035627 COSM32547 |
416 | I>V | a breast cancer sample; somatic mutation large_intestine breast [UniProt, Cosmic] | No |
ClinGen cosmic curated UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1342660219 CA412516503 |
421 | G>R | No |
ClinGen gnomAD |
|
|
CA412516223 rs1363181325 |
435 | V>L | No |
ClinGen TOPMed |
|
|
CA412516128 rs1406854160 |
441 | H>Y | No |
ClinGen gnomAD |
|
|
rs1569201390 CA412516100 |
443 | A>S | No |
ClinGen Ensembl |
|
|
rs1555931252 CA412516069 RCV000504430 |
445 | N>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412516021 rs1468374942 |
448 | F>L | No |
ClinGen TOPMed |
|
|
rs762253691 CA10366115 |
454 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10366114 rs754443220 |
456 | S>N | No |
ClinGen ExAC |
|
|
CA16621295 rs1064796388 RCV000481301 |
461 | T>A | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA10366113 rs764667081 |
479 | L>P | No |
ClinGen ExAC |
|
|
CA412515052 rs1343342088 |
480 | K>T | No |
ClinGen gnomAD |
|
|
COSM20565 rs1271090915 CA412515005 VAR_040629 |
483 | Y>C | a gastric adenocarcinoma sample; somatic mutation large_intestine stomach [UniProt, Cosmic] | No |
ClinGen cosmic curated UniProt TOPMed dbSNP |
|
RCV000627423 rs1555928736 |
488 | Y>missing | No |
ClinVar dbSNP |
|
|
CA327474370 rs937495958 |
495 | L>I | No |
ClinGen TOPMed gnomAD |
|
|
CA327474369 rs113475091 |
509 | K>R | No |
ClinGen Ensembl |
|
|
RCV000261508 rs886043293 |
515 | E>missing | No |
ClinVar dbSNP |
|
|
rs1220503066 CA412514768 |
517 | S>G | No |
ClinGen gnomAD |
|
|
CA412514755 rs1343122506 |
519 | V>I | No |
ClinGen gnomAD |
|
|
rs746538840 CA10366101 |
524 | T>I | No |
ClinGen ExAC |
|
|
CA412514701 rs1249392910 |
527 | V>A | No |
ClinGen TOPMed |
|
|
CA10366100 rs779360754 COSM457239 |
527 | V>I | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1183435589 COSM1119092 CA412514667 |
532 | A>T | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1057524314 RCV000444528 CA16609163 |
536 | V>F | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412514310 rs765149978 |
542 | P>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10366081 rs765149978 |
542 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1403980430 CA412514200 |
558 | R>Q | No |
ClinGen gnomAD |
|
|
rs1569194043 RCV000734392 CA412514189 |
560 | C>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA327474164 rs980055137 |
570 | A>V | No |
ClinGen TOPMed |
|
|
RCV000481699 rs1064795003 CA16621294 |
581 | T>A | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
RCV000422365 rs1057524393 CA16608385 |
600 | D>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1770398814 RCV001311821 |
609 | Y>F | No |
ClinVar dbSNP |
|
|
rs1413484122 CA412513726 |
611 | M>V | No |
ClinGen TOPMed |
|
|
rs1435847802 CA412513706 |
613 | T>I | No |
ClinGen gnomAD |
|
|
CA10366065 rs772004070 COSM182647 |
614 | G>S | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs755416120 CA10366055 |
622 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA10366054 rs751639594 |
623 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA10366053 rs150107747 RCV000366614 RCV001683162 |
628 | E>D | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA412513568 rs1210859060 |
632 | R>Q | No |
ClinGen TOPMed |
|
|
RCV000585468 rs1555926346 CA412513558 |
634 | G>C | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs758459388 CA10366052 |
635 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs750660360 CA10366051 |
636 | G>R | No |
ClinGen ExAC TOPMed |
|
|
rs1464801825 CA412513509 |
641 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
RCV000343124 rs886041328 CA10603612 |
645 | W>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412513451 rs1569190396 |
650 | D>H | No |
ClinGen Ensembl |
|
|
rs1009522729 CA327473550 |
658 | K>R | No |
ClinGen TOPMed |
|
|
rs1603417440 CA412512720 |
666 | Q>* | No |
ClinGen Ensembl |
|
|
CA10366038 rs769450157 |
669 | T>S | No |
ClinGen ExAC gnomAD |
|
|
COSM3844309 CA412512522 rs1285357297 |
680 | V>I | breast [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA412512092 rs1258505524 |
709 | A>S | No |
ClinGen gnomAD |
|
|
rs1323142539 CA412512053 |
712 | R>G | No |
ClinGen gnomAD |
|
|
CA327473502 rs1017565774 COSM1119082 |
712 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA327473501 VAR_040631 rs35026425 |
723 | R>C | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs759392909 CA10366027 |
731 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA412511759 rs1315964979 |
732 | I>V | No |
ClinGen gnomAD |
2 associated diseases with P51812
[MIM: 303600]: Coffin-Lowry syndrome (CLS)
An X-linked disorder characterized by intellectual disability associated with facial and digital dysmorphisms, progressive skeletal malformations, growth retardation, hearing deficit and paroxysmal movement disorders. {ECO:0000269|PubMed:10094187, ECO:0000269|PubMed:10528858, ECO:0000269|PubMed:14986828, ECO:0000269|PubMed:15214012, ECO:0000269|PubMed:8955270, ECO:0000269|PubMed:9837815}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 300844]: Intellectual developmental disorder, X-linked 19 (XLID19)
A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. {ECO:0000269|PubMed:10319851, ECO:0000269|PubMed:17100996}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An X-linked disorder characterized by intellectual disability associated with facial and digital dysmorphisms, progressive skeletal malformations, growth retardation, hearing deficit and paroxysmal movement disorders. {ECO:0000269|PubMed:10094187, ECO:0000269|PubMed:10528858, ECO:0000269|PubMed:14986828, ECO:0000269|PubMed:15214012, ECO:0000269|PubMed:8955270, ECO:0000269|PubMed:9837815}. Note=The disease is caused by variants affecting the gene represented in this entry.
- A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. {ECO:0000269|PubMed:10319851, ECO:0000269|PubMed:17100996}. Note=The disease is caused by variants affecting the gene represented in this entry.
10 regional properties for P51812
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 68 - 327 | IPR000719-1 |
| domain | Protein kinase domain | 422 - 679 | IPR000719-2 |
| domain | AGC-kinase, C-terminal | 328 - 397 | IPR000961 |
| active_site | Serine/threonine-protein kinase, active site | 189 - 201 | IPR008271-1 |
| active_site | Serine/threonine-protein kinase, active site | 535 - 547 | IPR008271-2 |
| binding_site | Protein kinase, ATP binding site | 74 - 100 | IPR017441-1 |
| binding_site | Protein kinase, ATP binding site | 428 - 451 | IPR017441-2 |
| domain | Protein kinase, C-terminal | 351 - 387 | IPR017892 |
| domain | Ribosomal protein S6 kinase alpha-3, C-terminal catalytic domain | 402 - 740 | IPR041905 |
| domain | Ribosomal S6 kinase, N-terminal catalytic domain | 72 - 388 | IPR041906 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24351 | RIBOSOMAL PROTEIN S6 KINASE |
| PANTHER Subfamily | PTHR24351:SF58 | RIBOSOMAL PROTEIN S6 KINASE ALPHA-3 |
| PANTHER Protein Class | protein modifying enzyme | |
| PANTHER Pathway Category |
Ras Pathway p90RSK Interleukin signaling pathway p90RSK Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade P90 RSK CCKR signaling map RSK1/2 PDGF signaling pathway p90RSK |
|
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| cysteine-type endopeptidase inhibitor activity involved in apoptotic process | Binds to and stops, prevents or reduces the activity of a cysteine-type endopeptidase involved in the apoptotic process. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| ribosomal protein S6 kinase activity | Catalysis of the reaction: ribosomal protein S6 + ATP = ribosomal protein S6 phosphate + ATP. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| chemical synaptic transmission | The vesicular release of classical neurotransmitter molecules from a presynapse, across a chemical synapse, the subsequent activation of neurotransmitter receptors at the postsynapse of a target cell (neuron, muscle, or secretory cell) and the effects of this activation on the postsynaptic membrane potential and ionic composition of the postsynaptic cytosol. This process encompasses both spontaneous and evoked release of neurotransmitter and all parts of synaptic vesicle exocytosis. Evoked transmission starts with the arrival of an action potential at the presynapse. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of a cysteine-type endopeptidase activity involved in the apoptotic process. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of cell differentiation | Any process that activates or increases the frequency, rate or extent of cell differentiation. |
| positive regulation of cell growth | Any process that activates or increases the frequency, rate, extent or direction of cell growth. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of DNA-templated transcription in response to stress | Modulation of the frequency, rate or extent of transcription from a DNA template as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation). |
| regulation of translation in response to stress | Modulation of the frequency, rate or extent of translation as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation). |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| skeletal system development | The process whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure. The skeleton is the bony framework of the body in vertebrates (endoskeleton) or the hard outer envelope of insects (exoskeleton or dermoskeleton). |
| toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor on the surface of a target cell. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P18652 | RPS6KA | Ribosomal protein S6 kinase 2 alpha | Gallus gallus (Chicken) | SS |
| Q5F3L1 | RPS6KA5 | Ribosomal protein S6 kinase alpha-5 | Gallus gallus (Chicken) | SS |
| Q96S38 | RPS6KC1 | Ribosomal protein S6 kinase delta-1 | Homo sapiens (Human) | PR |
| Q15349 | RPS6KA2 | Ribosomal protein S6 kinase alpha-2 | Homo sapiens (Human) | SS |
| Q15418 | RPS6KA1 | Ribosomal protein S6 kinase alpha-1 | Homo sapiens (Human) | EV |
| Q9UK32 | RPS6KA6 | Ribosomal protein S6 kinase alpha-6 | Homo sapiens (Human) | SS |
| O75676 | RPS6KA4 | Ribosomal protein S6 kinase alpha-4 | Homo sapiens (Human) | SS |
| O75582 | RPS6KA5 | Ribosomal protein S6 kinase alpha-5 | Homo sapiens (Human) | SS |
| Q9UBS0 | RPS6KB2 | Ribosomal protein S6 kinase beta-2 | Homo sapiens (Human) | PR |
| P23443 | RPS6KB1 | Ribosomal protein S6 kinase beta-1 | Homo sapiens (Human) | EV SS |
| Q9WUT3 | Rps6ka2 | Ribosomal protein S6 kinase alpha-2 | Mus musculus (Mouse) | SS |
| Q9Z2B9 | Rps6ka4 | Ribosomal protein S6 kinase alpha-4 | Mus musculus (Mouse) | SS |
| Q8C050 | Rps6ka5 | Ribosomal protein S6 kinase alpha-5 | Mus musculus (Mouse) | PR |
| Q8BLK9 | Rps6kc1 | Ribosomal protein S6 kinase delta-1 | Mus musculus (Mouse) | PR |
| P18653 | Rps6ka1 | Ribosomal protein S6 kinase alpha-1 | Mus musculus (Mouse) | SS |
| P18654 | Rps6ka3 | Ribosomal protein S6 kinase alpha-3 | Mus musculus (Mouse) | SS |
| Q63531 | Rps6ka1 | Ribosomal protein S6 kinase alpha-1 | Rattus norvegicus (Rat) | SS |
| Q18846 | rskn-2 | Putative ribosomal protein S6 kinase alpha-2 | Caenorhabditis elegans | PR |
| Q21734 | rskn-1 | Putative ribosomal protein S6 kinase alpha-1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPLAQLADPW | QKMAVESPSD | SAENGQQIMD | EPMGEEEINP | QTEEVSIKEI | AITHHVKEGH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EKADPSQFEL | LKVLGQGSFG | KVFLVKKISG | SDARQLYAMK | VLKKATLKVR | DRVRTKMERD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ILVEVNHPFI | VKLHYAFQTE | GKLYLILDFL | RGGDLFTRLS | KEVMFTEEDV | KFYLAELALA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LDHLHSLGII | YRDLKPENIL | LDEEGHIKLT | DFGLSKESID | HEKKAYSFCG | TVEYMAPEVV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NRRGHTQSAD | WWSFGVLMFE | MLTGTLPFQG | KDRKETMTMI | LKAKLGMPQF | LSPEAQSLLR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MLFKRNPANR | LGAGPDGVEE | IKRHSFFSTI | DWNKLYRREI | HPPFKPATGR | PEDTFYFDPE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FTAKTPKDSP | GIPPSANAHQ | LFRGFSFVAI | TSDDESQAMQ | TVGVHSIVQQ | LHRNSIQFTD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GYEVKEDIGV | GSYSVCKRCI | HKATNMEFAV | KIIDKSKRDP | TEEIEILLRY | GQHPNIITLK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DVYDDGKYVY | VVTELMKGGE | LLDKILRQKF | FSEREASAVL | FTITKTVEYL | HAQGVVHRDL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KPSNILYVDE | SGNPESIRIC | DFGFAKQLRA | ENGLLMTPCY | TANFVAPEVL | KRQGYDAACD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| IWSLGVLLYT | MLTGYTPFAN | GPDDTPEEIL | ARIGSGKFSL | SGGYWNSVSD | TAKDLVSKML |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HVDPHQRLTA | ALVLRHPWIV | HWDQLPQYQL | NRQDAPHLVK | GAMAATYSAL | NRNQSPVLEP |
| 730 | |||||
| VGRSTLAQRR | GIKKITSTAL |