P51809
Gene name |
VAMP7 (SYBL1) |
Protein name |
Vesicle-associated membrane protein 7 |
Names |
VAMP-7, Synaptobrevin-like protein 1, Tetanus-insensitive VAMP, Ti-VAMP |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6845 |
EC number |
|
Protein Class |
SNARE PROTEINS (PTHR21136) |
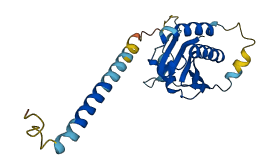
Descriptions
VAMP7 is involved in the targeting and/or fusion of transport vesicles to their target membrane. Longin domain negatively regulates the ability of VAMP7 like other SNARE attachment proteins including Ykt6.
Autoinhibitory domains (AIDs)
Target domain |
125-185 (v-SNARE coiled-coil homology) |
Relief mechanism |
|
Assay |
Deletion assay |
Accessory elements
No accessory elements
References
- Wen W et al. (2010) "Lipid-Induced conformational switch controls fusion activity of longin domain SNARE Ykt6", Molecular cell, 37, 383-95
- Martinez-Arca S et al. (2003) "A dual mechanism controlling the localization and function of exocytic v-SNAREs", Proceedings of the National Academy of Sciences of the United States of America, 100, 9011-6
- Gonzalez LC Jr et al. (2001) "A novel snare N-terminal domain revealed by the crystal structure of Sec22b", The Journal of biological chemistry, 276, 24203-11
- Mancias JD et al. (2007) "The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope", Molecular cell, 26, 403-14
Autoinhibited structure

Activated structure

2 structures for P51809
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DMW | NMR | - | A | 1-118 | PDB |
| AF-P51809-F1 | Predicted | AlphaFoldDB |
136 variants for P51809
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA10569808 rs749628563 |
2 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA414944841 rs1393254765 |
6 | A>S | No |
ClinGen TOPMed |
|
|
rs1374161238 CA414944871 |
9 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA414944869 rs1374161238 |
9 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA10569812 rs747707644 |
10 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA414944981 rs1292974855 |
14 | I>N | No |
ClinGen gnomAD |
|
|
rs373229104 CA10569815 |
14 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs770972520 CA10569816 |
16 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs776611068 CA10569817 |
17 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1009943478 CA337424754 |
19 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA414945096 rs764438604 |
19 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10569819 rs764438604 |
19 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337424758 rs1022322237 |
21 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA414945259 rs1485555278 |
25 | F>I | No |
ClinGen gnomAD |
|
|
CA414945350 rs1452475732 |
27 | E>D | No |
ClinGen gnomAD |
|
|
rs774375452 CA10569820 |
27 | E>V | No |
ClinGen ExAC gnomAD |
|
|
rs200207782 CA10569821 |
28 | V>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1423523366 CA414945388 |
29 | T>A | No |
ClinGen gnomAD |
|
|
CA414945573 rs1224322450 |
34 | A>D | No |
ClinGen TOPMed |
|
|
CA10569822 rs777066855 |
36 | I>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA10569825 rs766838352 |
44 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs1286683976 COSM278068 CA414945876 |
45 | Y>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs755283702 CA10569827 |
46 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA10569828 rs778633243 |
47 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs1242077651 CA414945946 |
49 | N>H | No |
ClinGen gnomAD |
|
|
CA10569842 rs766928340 |
55 | I>V | No |
ClinGen ExAC |
|
|
CA414947741 rs1277706821 |
58 | D>V | No |
ClinGen gnomAD |
|
|
CA414947764 rs1338300950 |
60 | I>T | No |
ClinGen gnomAD |
|
|
CA414947782 rs1270722806 |
63 | L>V | No |
ClinGen gnomAD |
|
|
CA414947809 rs1343983731 |
67 | D>N | No |
ClinGen gnomAD |
|
|
rs1397336378 CA414947819 |
68 | D>Y | No |
ClinGen TOPMed |
|
|
CA414947845 rs1383465000 |
70 | F>I | No |
ClinGen TOPMed |
|
|
CA414947854 rs1178045298 |
71 | E>Q | No |
ClinGen gnomAD |
|
|
CA10569865 rs763365685 |
72 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414947862 rs763365685 |
72 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763855733 CA10569866 |
72 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414947864 rs763855733 |
72 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337426786 rs1042066091 |
73 | S>F | No |
ClinGen TOPMed |
|
|
CA10569870 rs756855221 |
74 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756855221 CA10569869 |
74 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749923707 CA414947870 |
74 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749923707 CA10569871 |
74 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414947875 rs1221292959 |
75 | A>D | No |
ClinGen gnomAD |
|
|
rs142789451 CA10569873 |
77 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs142789451 CA10569872 |
77 | N>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs749093806 CA10569874 |
80 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA10569875 rs144784802 |
81 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA337426795 rs1054487918 |
82 | I>L | No |
ClinGen TOPMed |
|
|
rs778002287 CA10569877 |
82 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414947921 rs1569436332 |
82 | I>T | No |
ClinGen Ensembl |
|
|
rs1054487918 CA414947920 |
82 | I>V | No |
ClinGen TOPMed |
|
|
CA414947925 rs1272623789 |
83 | K>E | No |
ClinGen TOPMed |
|
|
rs1215934856 CA414947931 |
83 | K>N | No |
ClinGen gnomAD |
|
|
rs747186212 CA10569878 |
84 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA414947945 rs1484720485 |
85 | R>S | No |
ClinGen gnomAD |
|
|
rs771121932 CA10569879 |
86 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA10569880 rs776616389 |
88 | T>A | No |
ClinGen ExAC |
|
|
rs759659358 CA10569881 |
88 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770265952 CA10569882 |
89 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA414947976 rs147961723 |
90 | Y>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs764509692 CA10569885 |
95 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
COSM756723 rs1401961705 CA414948023 COSM1151556 |
98 | L>V | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs761603793 CA10569887 |
101 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767245575 CA10569888 |
102 | M>K | No |
ClinGen ExAC gnomAD |
|
|
CA10569889 rs749965834 |
104 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA414948070 rs1439223778 |
105 | E>K | No |
ClinGen gnomAD |
|
|
rs1312056711 CA414948093 |
108 | S>G | No |
ClinGen gnomAD |
|
|
rs748761504 CA337427183 |
115 | K>R | No |
ClinGen Ensembl |
|
|
CA414948868 rs1394193454 |
118 | S>F | No |
ClinGen TOPMed |
|
|
CA414948883 rs1169759773 |
119 | E>D | No |
ClinGen TOPMed |
|
|
CA414948891 rs1333789051 |
120 | N>D | No |
ClinGen gnomAD |
|
|
CA414948924 rs1259689536 |
122 | G>D | No |
ClinGen gnomAD |
|
|
CA414948922 rs189743661 |
122 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
RCV000979158 CA10569904 rs189743661 |
122 | G>S | No |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs1262967711 CA414948950 |
124 | D>E | No |
ClinGen gnomAD |
|
|
CA10569905 rs767131949 |
124 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs147108180 CA337427189 |
125 | K>E | No |
ClinGen ESP gnomAD |
|
|
CA10569906 rs772862018 |
126 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA414949005 rs1602936658 |
127 | M>I | No |
ClinGen Ensembl |
|
|
CA414948994 rs145477164 |
127 | M>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1231153270 CA414948987 |
127 | M>L | No |
ClinGen gnomAD |
|
|
RCV000895187 CA10569907 rs145477164 |
127 | M>T | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA10569908 rs374448760 |
130 | Q>E | No |
ClinGen ESP ExAC gnomAD |
|
|
CA414949148 rs1474314381 |
133 | V>G | No |
ClinGen gnomAD |
|
|
rs765156353 CA10569911 |
134 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs367823371 CA414949179 |
135 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10569913 rs148820711 |
137 | K>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1361452596 CA414949239 |
139 | I>S | No |
ClinGen gnomAD |
|
|
rs1293439191 CA414949256 |
140 | M>K | No |
ClinGen gnomAD |
|
|
rs781279551 CA10569914 |
140 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10569915 rs781279551 |
140 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10569917 rs780227295 |
141 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs756165763 CA10569916 |
141 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414949265 rs756165763 |
141 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414949303 rs1569437620 |
143 | N>S | No |
ClinGen Ensembl |
|
|
CA10569919 rs769350937 |
144 | I>R | No |
ClinGen ExAC gnomAD |
|
|
rs769350937 CA414949320 |
144 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs749343955 CA10569918 |
144 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1195050277 CA414949329 |
145 | D>N | No |
ClinGen gnomAD |
|
|
rs778240818 CA10569955 |
147 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs772345066 CA10569954 |
147 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747331110 CA10569956 |
150 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA414891910 rs1328107352 |
164 | L>F | No |
ClinGen TOPMed |
|
|
rs143522328 CA10569958 |
165 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs143522328 CA10569959 |
165 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs143522328 CA10569960 |
165 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414892006 rs1336206221 |
166 | D>E | No |
ClinGen TOPMed |
|
|
rs147995235 CA10569979 |
169 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414899844 rs1325046086 |
171 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1320346446 CA414900026 |
176 | R>S | No |
ClinGen TOPMed |
|
|
CA10569981 rs768699851 |
177 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10569980 rs768699851 |
177 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10569984 rs777015986 |
180 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs377088658 CA10569985 |
180 | R>Q | No |
ClinGen ESP ExAC gnomAD |
|
|
rs765552422 CA10569986 |
184 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs369451370 CA10569987 |
187 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs971184157 CA337350454 |
189 | L>F | No |
ClinGen TOPMed |
|
|
CA10569991 rs144740706 |
193 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414900591 rs1187710168 |
194 | I>S | No |
ClinGen TOPMed |
|
|
rs751968119 CA10569992 |
195 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs141889109 CA10569994 |
196 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs141889109 CA10569995 |
196 | V>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1419558765 CA414900673 |
198 | I>V | No |
ClinGen gnomAD |
|
|
CA10570024 rs746803961 |
199 | V>M | No |
ClinGen ExAC |
|
|
CA10570027 rs763130384 |
202 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1215512019 CA414901997 |
205 | V>F | No |
ClinGen gnomAD |
|
|
rs1358017182 CA414902027 |
206 | S>L | No |
ClinGen TOPMed |
|
|
rs150795669 CA337351574 |
207 | P>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA414902077 rs1306133232 |
208 | L>V | No |
ClinGen gnomAD |
|
|
CA414902106 rs1237097249 |
209 | C>G | No |
ClinGen gnomAD |
|
|
CA337351583 rs977115691 |
210 | G>C | No |
ClinGen TOPMed |
|
|
CA414902167 rs1281695553 |
211 | G>R | No |
ClinGen gnomAD |
|
|
CA414902320 rs1324136728 |
216 | S>N | No |
ClinGen TOPMed |
|
|
rs1347944995 CA414902360 |
218 | V>M | No |
ClinGen gnomAD |
|
|
CA10570028 rs768887885 |
220 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA414902421 rs1235381216 |
220 | K>T | No |
ClinGen gnomAD |
No associated diseases with P51809
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR21136 | SNARE PROTEINS |
| PANTHER Subfamily | PTHR21136:SF196 | VESICLE-ASSOCIATED MEMBRANE PROTEIN 7 |
| PANTHER Protein Class | SNARE protein | |
| PANTHER Pathway Category | No pathway information available | |
23 GO annotations of cellular component
| Name | Definition |
|---|---|
| azurophil granule membrane | The lipid bilayer surrounding an azurophil granule, a primary lysosomal granule found in neutrophil granulocytes that contains a wide range of hydrolytic enzymes and is released into the extracellular fluid. |
| clathrin-coated endocytic vesicle membrane | The lipid bilayer surrounding a clathrin-coated endocytic vesicle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| late endosome membrane | The lipid bilayer surrounding a late endosome. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| phagocytic vesicle | A membrane-bounded intracellular vesicle that arises from the ingestion of particulate material by phagocytosis. |
| phagocytic vesicle membrane | The lipid bilayer surrounding a phagocytic vesicle. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| platelet alpha granule | A secretory organelle found in blood platelets, which is unique in that it exhibits further compartmentalization and acquires its protein content via two distinct mechanisms: (1) biosynthesis predominantly at the megakaryocyte (MK) level (with some vestigial platelet synthesis) (e.g. platelet factor 4) and (2) endocytosis and pinocytosis at both the MK and circulating platelet levels (e.g. fibrinogen (Fg) and IgG). |
| pseudopodium | A temporary protrusion or retractile process of a cell, associated with flowing movements of the protoplasm, and serving for locomotion and feeding. |
| secretory granule | A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules. |
| secretory granule membrane | The lipid bilayer surrounding a secretory granule. |
| SNARE complex | A protein complex involved in membrane fusion; a stable ternary complex consisting of a four-helix bundle, usually formed from one R-SNARE and three Q-SNAREs with an ionic layer sandwiched between hydrophobic layers. One well-characterized example is the neuronal SNARE complex formed of synaptobrevin 2, syntaxin 1a, and SNAP-25. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
| trans-Golgi network | The network of interconnected tubular and cisternal structures located within the Golgi apparatus on the side distal to the endoplasmic reticulum, from which secretory vesicles emerge. The trans-Golgi network is important in the later stages of protein secretion where it is thought to play a key role in the sorting and targeting of secreted proteins to the correct destination. |
| transport vesicle membrane | The lipid bilayer surrounding a transport vesicle. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| SNAP receptor activity | Acting as a marker to identify a membrane and interacting selectively with one or more SNAREs on another membrane to mediate membrane fusion. |
| SNARE binding | Binding to a SNARE (soluble N-ethylmaleimide-sensitive factor attached protein receptor) protein. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| calcium-ion regulated exocytosis | The release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle by fusion of the vesicle with the plasma membrane of a cell, induced by a rise in cytosolic calcium-ion levels. |
| endoplasmic reticulum to Golgi vesicle-mediated transport | The directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi. |
| endosome to lysosome transport | The directed movement of substances from endosomes to lysosomes. |
| eosinophil degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as major basic protein, eosinophil peroxidase, and eosinophil cationic protein by an eosinophil. |
| exocytosis | A process of secretion by a cell that results in the release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle. Exocytosis can occur either by full fusion, when the vesicle collapses into the plasma membrane, or by a kiss-and-run mechanism that involves the formation of a transient contact, a pore, between a granule (for exemple of chromaffin cells) and the plasma membrane. The latter process most of the time leads to only partial secretion of the granule content. Exocytosis begins with steps that prepare vesicles for fusion with the membrane (tethering and docking) and ends when molecules are secreted from the cell. |
| natural killer cell degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as perforin and granzymes by a natural killer cell. |
| neutrophil degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as proteases, lipases, and inflammatory mediators by a neutrophil. |
| phagocytosis, engulfment | The internalization of bacteria, immune complexes and other particulate matter or of an apoptotic cell by phagocytosis, including the membrane and cytoskeletal processes required, which involves one of three mechanisms: zippering of pseudopods around a target via repeated receptor-ligand interactions, sinking of the target directly into plasma membrane of the phagocytosing cell, or induced uptake via an enhanced membrane ruffling of the phagocytosing cell similar to macropinocytosis. |
| positive regulation of histamine secretion by mast cell | Any process that activates or increases the frequency, rate or extent of histamine secretion by mast cell. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| vesicle fusion | Fusion of the membrane of a transport vesicle with its target membrane. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q12255 | NYV1 | Vacuolar v-SNARE NYV1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q17QI5 | VAMP7 | Vesicle-associated membrane protein 7 | Bos taurus (Bovine) | SS |
| Q5ZL74 | VAMP7 | Vesicle-associated membrane protein 7 | Gallus gallus (Chicken) | SS |
| P70280 | Vamp7 | Vesicle-associated membrane protein 7 | Mus musculus (Mouse) | SS |
| Q9JHW5 | Vamp7 | Vesicle-associated membrane protein 7 | Rattus norvegicus (Rat) | SS |
| Q9M376 | VAMP727 | Vesicle-associated membrane protein 727 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LFP1 | VAMP713 | Vesicle-associated membrane protein 713 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SIQ9 | VAMP712 | Vesicle-associated membrane protein 712 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49377 | VAMP711 | Vesicle-associated membrane protein 711 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FMR5 | VAMP714 | Vesicle-associated membrane protein 714 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAILFAVVAR | GTTILAKHAW | CGGNFLEVTE | QILAKIPSEN | NKLTYSHGNY | LFHYICQDRI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VYLCITDDDF | ERSRAFNFLN | EIKKRFQTTY | GSRAQTALPY | AMNSEFSSVL | AAQLKHHSEN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KGLDKVMETQ | AQVDELKGIM | VRNIDLVAQR | GERLELLIDK | TENLVDSSVT | FKTTSRNLAR |
| 190 | 200 | 210 | |||
| AMCMKNLKLT | IIIIIVSIVF | IYIIVSPLCG | GFTWPSCVKK |