P51159
Gene name |
RAB27A (RAB27) |
Protein name |
Ras-related protein Rab-27A |
Names |
Rab-27, GTP-binding protein Ram |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5873 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
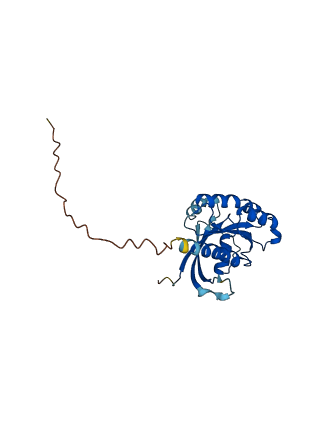
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for P51159
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6HUF | X-ray | 282 A | A/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P | 2-192 | PDB |
| 7OPP | X-ray | 232 A | A/C | 1-192 | PDB |
| 7OPQ | X-ray | 223 A | A/B | 1-192 | PDB |
| 7OPR | X-ray | 232 A | A/B | 1-192 | PDB |
| AF-P51159-F1 | Predicted | AlphaFoldDB |
213 variants for P51159
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs141281020 RCV000501657 RCV002263710 |
1 | M>T | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA7573715 RCV002263986 RCV000795519 rs539575657 |
4 | G>V | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs1555394745 RCV000644917 CA618059762 RCV000521818 |
6 | Y>* | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs145253993 RCV000644920 RCV002263884 CA7573714 |
6 | Y>C | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs1595700039 RCV000006354 |
18 | S>missing | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs778448317 RCV000793188 CA7573706 |
32 | G>R | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001860238 rs770601673 RCV000606194 |
50 | R>missing | Griscelli syndrome type 2 Griscelli syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA7573675 RCV002262993 RCV000551972 rs540520068 |
56 | S>N | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
CA7573673 RCV001246273 RCV002264250 RCV002568649 rs201284258 |
58 | P>S | Griscelli syndrome type 2 Autoinflammatory syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs770949071 CA7573670 RCV001217202 |
59 | D>H | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1895985830 RCV001217895 |
63 | G>C | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV002264276 rs1371360853 CA392530622 RCV001351363 |
70 | L>P | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs776920896 CA7573665 CA7573664 RCV001064342 |
71 | Q>H | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
VAR_010654 CA117886 rs28938176 RCV000006348 |
73 | W>G | Griscelli syndrome type 2 GS2; does not affect GTP binding; cannot interact with MLPH; significant reduction in interaction with UNC13D; abolishes localization to lysosomes [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA7573638 RCV000483557 RCV000850516 rs753966933 RCV002263695 |
82 | R>C | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs140640177 RCV001316203 CA7573637 |
82 | R>H | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC dbSNP gnomAD |
|
RCV001215059 rs1895911610 |
84 | L>missing | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA7573630 RCV001317455 rs369777755 |
86 | T>I | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA117898 RCV000169674 rs104894497 RCV000524022 RCV000006357 |
87 | A>P | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA7573625 rs137960099 RCV002263930 RCV000685947 |
92 | A>V | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001116408 CA7573622 RCV002264191 rs144492641 RCV002558151 |
103 | N>D | Griscelli syndrome type 2 Autoinflammatory syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001939967 rs756644243 |
112 | N>missing | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA117895 RCV001778648 rs104894500 RCV000006356 RCV002251884 |
118 | Q>* | Griscelli syndrome type 2 Multisystem inflammatory syndrome in children [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
CA392529856 RCV000644916 rs1259290779 |
122 | Y>F | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs1895696380 RCV001222124 RCV002562548 |
126 | P>missing | Griscelli syndrome type 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA7573585 RCV001211511 rs143719577 |
128 | I>T | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_011334 rs104894498 RCV000006352 CA117889 |
130 | L>P | Griscelli syndrome type 2 GS2; strongly affects GTP binding; cannot interact with MLPH [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000644919 rs150463407 RCV001354874 CA7573579 RCV002263883 |
140 | Q>E | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs766575263 RCV000793635 |
141 | R>missing | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA7573575 RCV001061386 rs569221128 |
142 | V>A | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes dbSNP |
|
rs538323738 RCV001041482 RCV002551497 CA7573577 |
142 | V>L | Griscelli syndrome type 2 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV000006353 rs104894499 VAR_011335 CA117892 |
152 | A>P | Griscelli syndrome type 2 GS2; interferes with melanosome transport [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
|
CA7573570 rs200031368 RCV001242800 |
156 | G>A | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs746081723 CA7573551 RCV001056261 |
158 | P>R | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001251148 rs1894614852 |
159 | Y>H | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001232890 rs1894613403 |
166 | N>K | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001053198 CA7573549 rs576324438 |
166 | N>S | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001090830 rs767481076 RCV000477784 |
172 | Q>missing | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs142217102 RCV000823371 CA7573545 |
173 | A>G | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs751121802 RCV001205753 CA7573543 |
174 | I>V | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA7573542 RCV000697369 rs757760608 |
176 | M>L | Griscelli syndrome type 2 Variant assessed as Somatic; 0.0 impact. [ClinVar, NCI-TCGA] | Yes |
ClinGen ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA7573536 rs139025012 RCV001796243 RCV002264003 RCV000811106 |
181 | I>M | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001090829 RCV000499859 RCV002263709 CA7573534 rs200956636 |
184 | R>* | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs141362723 CA7573533 RCV000700237 RCV002263944 |
184 | R>Q | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000224304 CA7573530 rs182849552 RCV001027844 RCV002262831 |
187 | R>Q | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002264024 rs144946000 CA7573531 RCV000823302 |
187 | R>W | Griscelli syndrome type 2 Autoinflammatory syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002543819 RCV001321106 CA7573526 rs554967424 |
198 | V>A | Griscelli syndrome type 2 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs778390086 RCV001343347 CA392781820 |
199 | V>A | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV001056643 rs928880499 CA271242129 |
205 | A>S | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV000696747 rs151048993 CA7573519 |
207 | T>M | Griscelli syndrome type 2 Variant assessed as Somatic; 0.0 impact. [ClinVar, NCI-TCGA] | Yes |
ClinGen ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV001064343 CA7573512 rs373190916 |
218 | A>T | Griscelli syndrome type 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA392531179 rs1220538156 |
4 | G>R | No |
ClinGen gnomAD |
|
|
CA270737389 rs952920589 |
5 | D>E | No |
ClinGen TOPMed |
|
| TCGA novel | 5 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs749531175 CA7573713 |
7 | D>G | No |
ClinGen ExAC |
|
|
CA392531145 rs1253221848 |
7 | D>H | No |
ClinGen TOPMed |
|
| TCGA novel | 12 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1001351367 CA270737356 |
12 | F>S | No |
ClinGen TOPMed gnomAD |
|
|
CA270737353 rs905779372 |
13 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA270737346 rs1024213806 |
14 | A>V | No |
ClinGen Ensembl |
|
|
CA392531034 rs1398927780 |
16 | G>R | No |
ClinGen gnomAD |
|
|
rs777995262 CA7573712 |
17 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA392530993 rs1466620676 |
19 | G>C | No |
ClinGen TOPMed gnomAD |
|
|
rs768784784 CA270737330 |
19 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA7573711 rs369478092 |
20 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs369478092 CA392530985 |
20 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1200907732 CA392530962 |
22 | K>R | No |
ClinGen Ensembl |
|
|
CA392530952 rs1461262502 |
23 | T>S | No |
ClinGen TOPMed |
|
|
rs746770347 CA7573710 |
24 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1445499017 CA392530935 |
25 | V>I | No |
ClinGen gnomAD |
|
|
CA7573709 rs139145499 |
29 | Y>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs998047728 CA392530888 |
31 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA7573707 rs150782434 |
31 | D>G | No |
ClinGen ESP ExAC gnomAD |
|
|
CA7573705 rs756730276 |
33 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1332723274 CA392530879 |
33 | K>R | No |
ClinGen gnomAD |
|
|
rs1595699929 CA392530850 |
37 | K>T | No |
ClinGen Ensembl |
|
|
CA392530809 rs1416818050 |
43 | G>D | No |
ClinGen TOPMed |
|
|
rs763762181 CA7573703 |
44 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1131388 CA7573702 |
48 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1323893818 CA392530768 |
49 | K>* | No |
ClinGen gnomAD |
|
|
CA7573679 rs766872265 |
52 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA7573678 rs762968655 |
53 | Y>D | No |
ClinGen ExAC gnomAD |
|
|
CA392530727 rs1288513868 |
53 | Y>F | No |
ClinGen gnomAD |
|
|
rs201878183 CA7573671 |
58 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs201878183 CA7573672 |
58 | P>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7573669 VAR_028206 rs1050930 |
62 | T>S | No |
ClinGen UniProt ExAC TOPMed dbSNP gnomAD |
|
|
CA7573668 rs777409501 |
64 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA7573667 rs755674449 |
65 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA270734172 rs762691442 |
67 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs762691442 CA7573666 |
67 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs1173348917 CA392530635 |
68 | I>M | No |
ClinGen gnomAD |
|
|
rs1259068708 CA392530626 |
70 | L>M | No |
ClinGen TOPMed |
|
|
CA7573663 rs751907735 |
72 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA392530586 rs1489981016 |
76 | A>P | No |
ClinGen TOPMed |
|
|
rs1489981016 CA392530587 |
76 | A>T | No |
ClinGen TOPMed |
|
|
CA7573662 rs766632505 |
76 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7573660 rs750877483 |
77 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs758986523 CA7573661 |
77 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA392530576 rs1200627507 |
78 | Q>* | No |
ClinGen gnomAD |
|
|
rs765369750 CA7573659 |
80 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs140640177 CA7573636 |
82 | R>L | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
|
CA7573635 rs752851113 |
83 | S>C | No |
ClinGen ExAC gnomAD |
|
|
VAR_028207 rs4340274 CA270732861 |
84 | L>F | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP |
|
|
COSM963022 CA7573632 rs773233108 |
85 | T>M | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs719705 CA270732860 VAR_028208 |
85 | T>P | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs104894497 CA7573629 |
87 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7573628 rs201697259 |
87 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed |
|
|
rs905528188 CA270732854 |
89 | F>V | No |
ClinGen Ensembl |
|
|
CA392530485 rs1229857604 |
91 | D>Y | No |
ClinGen gnomAD |
|
|
CA7573626 rs772514326 |
92 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1379213338 CA392530475 |
93 | M>L | No |
ClinGen gnomAD |
|
|
rs1291929786 CA392530471 |
93 | M>T | No |
ClinGen gnomAD |
|
|
rs1379213338 CA392530474 |
93 | M>V | No |
ClinGen gnomAD |
|
|
rs757798952 CA7573623 |
94 | G>D | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 96 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs891946275 CA270732812 |
98 | L>V | No |
ClinGen gnomAD |
|
|
CA392530435 rs1371541124 |
99 | F>V | No |
ClinGen gnomAD |
|
|
rs1418292027 CA392530429 |
100 | D>H | No |
ClinGen gnomAD |
|
|
rs144492641 CA392530410 |
103 | N>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 104 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756090505 CA7573620 |
104 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA7573619 rs200107421 |
105 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7573618 rs200107421 |
105 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762830892 CA7573617 |
106 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA392530378 rs1477388225 |
107 | F>C | No |
ClinGen gnomAD |
|
|
rs765096212 CA7573615 |
109 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs149282056 CA7573616 |
109 | N>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs761912458 CA7573614 |
110 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs761912458 CA392530363 |
110 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1284959303 CA392530352 |
111 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1595693254 RCV000788137 |
112 | N>missing | No |
ClinVar dbSNP |
|
|
CA7573613 rs776700493 |
112 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA392530337 rs1299441468 |
113 | W>C | No |
ClinGen gnomAD |
|
|
rs1218762595 CA392530334 |
114 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs183582444 CA7573587 |
115 | S>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs183582444 CA270729457 |
115 | S>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs768213316 CA7573611 |
115 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs104894500 CA392529915 |
118 | Q>K | No |
ClinGen gnomAD |
|
|
CA392529888 rs1317327419 |
119 | M>I | No |
ClinGen gnomAD |
|
|
rs771291441 CA7573586 |
119 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771291441 CA270729443 |
119 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 120 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA392529858 rs1259290779 |
122 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1220936730 CA392529850 |
123 | C>G | No |
ClinGen gnomAD |
|
|
CA270729438 rs755454272 |
124 | E>K | No |
ClinGen gnomAD |
|
|
rs1219246621 CA392529808 |
126 | P>L | No |
ClinGen TOPMed |
|
|
rs1343658287 CA392529815 |
126 | P>S | No |
ClinGen TOPMed |
|
|
rs1338258435 CA392529748 |
132 | G>V | No |
ClinGen gnomAD |
|
|
rs558000887 CA7573581 |
135 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA392529655 rs150463407 |
140 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA392529631 rs538323738 |
142 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1410069952 CA392529592 |
145 | E>Q | No |
ClinGen TOPMed |
|
|
CA7573574 rs780379424 |
145 | E>V | No |
ClinGen ExAC gnomAD |
|
|
rs373591014 CA270729387 |
147 | E>G | No |
ClinGen ESP TOPMed |
|
|
CA270729391 rs75258234 |
147 | E>K | No |
ClinGen gnomAD |
|
|
rs1238797403 CA392529557 |
148 | A>P | No |
ClinGen gnomAD |
|
|
CA392529545 COSM1517413 rs1197622697 |
149 | I>V | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs104894499 CA392529511 |
152 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 153 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1316745675 CA392529463 |
155 | Y>* | No |
ClinGen gnomAD |
|
|
rs1323839207 CA392529465 |
155 | Y>C | No |
ClinGen gnomAD |
|
|
rs375379384 CA271242137 |
157 | I>V | No |
ClinGen ESP TOPMed |
|
|
rs1351114357 CA392782081 |
159 | Y>C | No |
ClinGen Ensembl |
|
|
rs1411113252 CA392782054 |
163 | S>G | No |
ClinGen TOPMed |
|
|
CA392782051 rs1361456018 |
163 | S>T | No |
ClinGen gnomAD |
|
|
rs1017422312 CA271242135 |
165 | A>D | No |
ClinGen Ensembl |
|
|
CA271242134 rs960789100 |
167 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1377085164 CA392782008 |
170 | I>V | No |
ClinGen gnomAD |
|
|
rs1338691454 CA392782000 |
171 | S>G | No |
ClinGen TOPMed |
|
| TCGA novel | 173 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs752581278 CA7573547 |
173 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA7573546 rs142217102 |
173 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1488806292 CA392781971 |
175 | E>D | No |
ClinGen gnomAD |
|
|
rs1217489548 CA392781976 |
175 | E>K | No |
ClinGen TOPMed |
|
|
CA7573541 rs762664337 |
176 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs765537875 CA392781944 |
179 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA7573537 rs762174469 |
181 | I>K | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 182 | M>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA392781903 rs772264224 |
186 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA7573532 rs772264224 |
186 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1422163597 CA392781890 |
188 | C>Y | No |
ClinGen gnomAD |
|
|
rs769829050 CA392781885 |
189 | V>L | No |
ClinGen TOPMed |
|
|
CA271242133 rs769829050 |
189 | V>M | No |
ClinGen TOPMed |
|
|
CA7573529 rs373846556 |
190 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs747927730 CA7573528 |
190 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs780829484 CA7573527 |
192 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs865855583 CA271242132 |
195 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA392781830 rs1595661755 |
197 | G>E | No |
ClinGen Ensembl |
|
| TCGA novel | 197 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778390086 CA271242131 |
199 | V>E | No |
ClinGen gnomAD |
|
|
rs779599183 CA7573524 |
199 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs757849893 CA7573523 |
200 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA271242130 rs938803566 |
200 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| TCGA novel | 201 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7573522 rs758375931 |
202 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs764890991 CA7573521 |
203 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA7573520 rs761345593 |
204 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs928880499 CA392781790 |
205 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1434577440 CA392781781 |
206 | S>C | No |
ClinGen TOPMed |
|
|
CA271242128 rs977631664 |
207 | T>A | No |
ClinGen Ensembl |
|
|
rs776080472 CA7573516 |
209 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7573517 rs761321376 |
209 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA271242127 rs370554230 |
211 | S>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA392781741 rs1210017547 |
212 | E>D | No |
ClinGen gnomAD |
|
|
CA7573515 rs772282429 |
213 | E>G | No |
ClinGen ExAC gnomAD |
|
|
COSM328989 rs1466660727 CA392781725 |
214 | K>N | Variant assessed as Somatic; 0.0 impact. liver [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1472655424 CA392781706 |
217 | G>A | No |
ClinGen gnomAD |
|
|
CA7573513 rs759620500 |
217 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1183770943 CA392781697 |
219 | C>R | No |
ClinGen gnomAD |
|
|
CA7573511 rs771235265 |
219 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs749441289 CA7573510 |
220 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA271242126 rs80288229 |
221 | C>W | No |
ClinGen Ensembl |
|
|
CA392781683 rs1197456186 |
221 | C>Y | No |
ClinGen gnomAD |
1 associated diseases with P51159
[MIM: 607624]: Griscelli syndrome 2 (GS2)
Rare autosomal recessive disorder that results in pigmentary dilution of the skin and hair, the presence of large clumps of pigment in hair shafts, and an accumulation of melanosomes in melanocytes. GS2 patients also develop an uncontrolled T-lymphocyte and macrophage activation syndrome, known as hemophagocytic syndrome, leading to death in the absence of bone marrow transplantation. Neurological impairment is present in some patients, likely as a result of hemophagocytic syndrome. {ECO:0000269|PubMed:10835631, ECO:0000269|PubMed:12446441, ECO:0000269|PubMed:12531900, ECO:0000269|PubMed:15548590}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- Rare autosomal recessive disorder that results in pigmentary dilution of the skin and hair, the presence of large clumps of pigment in hair shafts, and an accumulation of melanosomes in melanocytes. GS2 patients also develop an uncontrolled T-lymphocyte and macrophage activation syndrome, known as hemophagocytic syndrome, leading to death in the absence of bone marrow transplantation. Neurological impairment is present in some patients, likely as a result of hemophagocytic syndrome. {ECO:0000269|PubMed:10835631, ECO:0000269|PubMed:12446441, ECO:0000269|PubMed:12531900, ECO:0000269|PubMed:15548590}. Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P51159
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 9 - 177 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
17 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| exocytic vesicle | A transport vesicle that mediates transport from an intracellular compartment to the plasma membrane, and fuses with the plasma membrane to release various cargo molecules, such as proteins or hormones, by exocytosis. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| late endosome | A prelysosomal endocytic organelle differentiated from early endosomes by lower lumenal pH and different protein composition. Late endosomes are more spherical than early endosomes and are mostly juxtanuclear, being concentrated near the microtubule organizing center. |
| lysosome | A small lytic vacuole that has cell cycle-independent morphology found in most animal cells and that contains a variety of hydrolases, most of which have their maximal activities in the pH range 5-6. The contained enzymes display latency if properly isolated. About 40 different lysosomal hydrolases are known and lysosomes have a great variety of morphologies and functions. |
| melanosome | A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells. |
| melanosome membrane | The lipid bilayer surrounding a melanosome. |
| multivesicular body membrane | The lipid bilayer surrounding a multivesicular body. |
| photoreceptor outer segment | The outer segment of a vertebrate photoreceptor that contains a stack of membrane discs embedded with photoreceptor proteins. |
| secretory granule | A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules. |
| secretory granule membrane | The lipid bilayer surrounding a secretory granule. |
| specific granule lumen | The volume enclosed by the membrane of a specific granule, a granule with a membranous, tubular internal structure, found primarily in mature neutrophil cells. Most are released into the extracellular fluid. Specific granules contain lactoferrin, lysozyme, vitamin B12 binding protein and elastase. |
| Weibel-Palade body | A large, elongated, rod-shaped secretory granule characteristic of vascular endothelial cells that contain a number of structurally and functionally distinct proteins, of which the best characterized are von Willebrand factor (VWF) and P-selectin. Weibel-Palade bodies are formed from the trans-Golgi network in a process that depends on VWF, which is densely packed in a highly organized manner, and on coat proteins that remain associated with the granules. Upon cell stimulation, regulated exocytosis releases the contained proteins to the cell surface, where they act in the recruitment of platelets and leukocytes and in inflammatory and vasoactive responses. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein activity | A molecular function regulator that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular processes. Intrinsic GTPase activity returns the G protein to its GDP-bound state. The return to the GDP-bound state can be accelerated by the action of a GTPase-activating protein (GAP). |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| myosin V binding | Binding to a class V myosin; myosin V is a dimeric molecule involved in intracellular transport. |
| protein domain specific binding | Binding to a specific domain of a protein. |
20 GO annotations of biological process
| Name | Definition |
|---|---|
| antigen processing and presentation | The process in which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex. |
| blood coagulation | The sequential process in which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers. |
| complement-dependent cytotoxicity | Cell killing caused by the membrane attack complex formed following complement activation. |
| cytotoxic T cell degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as perforin and granzymes by a cytotoxic T cell. |
| exocytosis | A process of secretion by a cell that results in the release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle. Exocytosis can occur either by full fusion, when the vesicle collapses into the plasma membrane, or by a kiss-and-run mechanism that involves the formation of a transient contact, a pore, between a granule (for exemple of chromaffin cells) and the plasma membrane. The latter process most of the time leads to only partial secretion of the granule content. Exocytosis begins with steps that prepare vesicles for fusion with the membrane (tethering and docking) and ends when molecules are secreted from the cell. |
| exosomal secretion | The process whereby a membrane-bounded vesicle is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. |
| melanocyte differentiation | The process in which a relatively unspecialized cell acquires specialized features of a melanocyte. |
| melanosome localization | Any process in which a melanosome is transported to, and/or maintained in, a specific location within the cell. |
| melanosome transport | The directed movement of melanosomes into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| multivesicular body organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a multivesicular body. A multivesicular body is a type of late endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body sorting pathway | A vesicle-mediated transport process in which transmembrane proteins are ubiquitylated to facilitate their entry into luminal vesicles of multivesicular bodies (MVBs); upon subsequent fusion of MVBs with lysosomes or vacuoles, the cargo proteins are degraded. |
| natural killer cell degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as perforin and granzymes by a natural killer cell. |
| positive regulation of constitutive secretory pathway | Any process that activates or increases the frequency, rate or extent of constitutive secretory pathway. |
| positive regulation of exocytosis | Any process that activates or increases the frequency, rate or extent of exocytosis. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of phagocytosis | Any process that activates or increases the frequency, rate or extent of phagocytosis. |
| positive regulation of reactive oxygen species biosynthetic process | Any process that activates or increases the frequency, rate or extent of reactive oxygen species biosynthetic process. |
| positive regulation of regulated secretory pathway | Any process that activates or increases the frequency, rate or extent of regulated secretory pathway. |
| synaptic vesicle transport | The directed movement of synaptic vesicles. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8HZJ5 | RAB27B | Ras-related protein Rab-27B | Bos taurus (Bovine) | PR |
| Q1HE58 | RAB27A | Ras-related protein Rab-27A | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| Q15771 | RAB30 | Ras-related protein Rab-30 | Homo sapiens (Human) | PR |
| O00194 | RAB27B | Ras-related protein Rab-27B | Homo sapiens (Human) | PR |
| Q9BZG1 | RAB34 | Ras-related protein Rab-34 | Homo sapiens (Human) | PR |
| O95755 | RAB36 | Ras-related protein Rab-36 | Homo sapiens (Human) | PR |
| Q99P58 | Rab27b | Ras-related protein Rab-27B | Mus musculus (Mouse) | PR |
| Q9ERI2 | Rab27a | Ras-related protein Rab-27A | Mus musculus (Mouse) | PR |
| Q4LE85 | RAB27A | Ras-related protein Rab-27A | Sus scrofa (Pig) | PR |
| Q99P74 | Rab27b | Ras-related protein Rab-27B | Rattus norvegicus (Rat) | PR |
| P23640 | Rab27a | Ras-related protein Rab-27A | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSDGDYDYLI | KFLALGDSGV | GKTSVLYQYT | DGKFNSKFIT | TVGIDFREKR | VVYRASGPDG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ATGRGQRIHL | QLWDTAGQER | FRSLTTAFFR | DAMGFLLLFD | LTNEQSFLNV | RNWISQLQMH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AYCENPDIVL | CGNKSDLEDQ | RVVKEEEAIA | LAEKYGIPYF | ETSAANGTNI | SQAIEMLLDL |
| 190 | 200 | 210 | 220 | ||
| IMKRMERCVD | KSWIPEGVVR | SNGHASTDQL | SEEKEKGACG | C |