P50750
Gene name |
CDK9 |
Protein name |
Cyclin-dependent kinase 9 |
Names |
C-2K, Cell division cycle 2-like protein kinase 4, Cell division protein kinase 9, Serine/threonine-protein kinase PITALRE, Tat-associated kinase complex catalytic subunit |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1025 |
EC number |
2.7.11.22: Protein-serine/threonine kinases |
Protein Class |
|
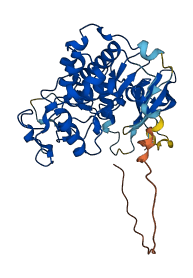
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
166-193 (Activation loop from InterPro)
Target domain |
19-315 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

29 structures for P50750
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3BLH | X-ray | 248 A | A | 2-330 | PDB |
| 3BLQ | X-ray | 290 A | A | 2-330 | PDB |
| 3BLR | X-ray | 280 A | A | 2-330 | PDB |
| 3LQ5 | X-ray | 300 A | A | 2-330 | PDB |
| 3MI9 | X-ray | 210 A | A | 1-345 | PDB |
| 3MIA | X-ray | 300 A | A | 1-345 | PDB |
| 3MY1 | X-ray | 280 A | A | 2-330 | PDB |
| 3TN8 | X-ray | 295 A | A | 2-330 | PDB |
| 3TNH | X-ray | 320 A | A | 2-330 | PDB |
| 3TNI | X-ray | 323 A | A | 2-330 | PDB |
| 4BCF | X-ray | 301 A | A | 2-330 | PDB |
| 4BCG | X-ray | 308 A | A | 2-330 | PDB |
| 4BCH | X-ray | 296 A | A | 2-330 | PDB |
| 4BCI | X-ray | 310 A | A | 2-330 | PDB |
| 4BCJ | X-ray | 316 A | A | 2-330 | PDB |
| 4EC8 | X-ray | 360 A | A | 2-372 | PDB |
| 4EC9 | X-ray | 321 A | A | 2-372 | PDB |
| 4IMY | X-ray | 294 A | A/C/E | 1-330 | PDB |
| 4OGR | X-ray | 300 A | A/E/I | 1-330 | PDB |
| 4OR5 | X-ray | 290 A | A/F | 7-332 | PDB |
| 5L1Z | X-ray | 590 A | A | 1-330 | PDB |
| 6CYT | X-ray | 350 A | A | 1-330 | PDB |
| 6GZH | X-ray | 317 A | A | 1-326 | PDB |
| 6W9E | X-ray | 310 A | A | 1-330 | PDB |
| 6Z45 | X-ray | 337 A | A | 1-330 | PDB |
| 7NWK | X-ray | 281 A | A | 1-330 | PDB |
| 8I0L | X-ray | 360 A | A | 2-330 | PDB |
| 8K5R | X-ray | 375 A | A | 1-330 | PDB |
| AF-P50750-F1 | Predicted | AlphaFoldDB |
252 variants for P50750
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA5251564 rs758230646 |
2 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA374994729 rs1290827114 |
2 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 3 | K>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374994760 rs1239571832 |
4 | Q>E | No |
ClinGen TOPMed |
|
|
rs777792275 CA5251565 |
5 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
CA374994780 rs1231461003 |
5 | Y>H | No |
ClinGen gnomAD |
|
|
COSM1187819 rs746864836 CA5251566 |
7 | S>L | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA200314040 rs566144307 |
9 | E>A | No |
ClinGen Ensembl |
|
|
rs1251086320 CA374994864 |
10 | C>G | No |
ClinGen gnomAD |
|
|
rs1456763070 CA374994892 |
11 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1456763070 CA374994898 |
11 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs781197980 CA5251568 |
17 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA374995041 rs1378710886 |
19 | Y>* | No |
ClinGen TOPMed |
|
|
rs770049111 CA5251570 |
22 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374995091 rs770049111 |
22 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374995101 rs1427479508 |
23 | A>T | No |
ClinGen gnomAD |
|
|
CA200314047 rs146765767 |
24 | K>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1243073913 CA374995402 |
32 | E>K | No |
ClinGen gnomAD |
|
|
rs1588537476 CA374995434 |
33 | V>G | No |
ClinGen Ensembl |
|
|
CA374995426 rs1474729591 |
33 | V>L | No |
ClinGen TOPMed |
|
|
CA374995486 rs779167622 |
36 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5251612 rs779167622 |
36 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs988782815 CA200314381 |
37 | R>G | No |
ClinGen TOPMed |
|
|
rs1283944680 CA374995534 |
39 | R>C | No |
ClinGen gnomAD |
|
|
CA200314385 rs914635422 |
39 | R>P | No |
ClinGen TOPMed |
|
|
rs1292765835 CA374995546 |
40 | K>Q | No |
ClinGen gnomAD |
|
|
rs905857521 CA200314388 |
41 | T>S | No |
ClinGen Ensembl |
|
|
CA5251616 rs747453571 |
42 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1588537494 CA374995646 |
45 | V>G | No |
ClinGen Ensembl |
|
|
rs761040673 CA200314396 |
50 | V>G | No |
ClinGen gnomAD |
|
| TCGA novel | 52 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374995704 rs1375660584 |
54 | N>H | No |
ClinGen Ensembl |
|
| TCGA novel | 54 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs149950953 CA5251620 |
58 | G>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA5251621 rs149950953 |
58 | G>W | No |
ClinGen ESP ExAC gnomAD |
|
|
VAR_041982 CA5251652 rs55640715 |
59 | F>L | No |
ClinGen UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1466578290 CA374996975 |
60 | P>R | No |
ClinGen gnomAD |
|
|
CA374996986 rs1293514453 |
61 | I>V | No |
ClinGen gnomAD |
|
|
rs376698069 CA5251655 |
64 | L>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA200314791 rs1014267439 |
65 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA374997020 rs1340146157 |
65 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA5251656 rs752013238 |
70 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1243972549 CA374997083 |
72 | L>F | No |
ClinGen gnomAD |
|
|
CA5251658 rs781568641 |
76 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA200314797 rs897679803 |
80 | N>H | No |
ClinGen Ensembl |
|
|
rs746324387 CA5251659 |
80 | N>K | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 80 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1426209808 CA374997171 |
81 | L>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA200314799 rs993418398 |
82 | I>T | No |
ClinGen gnomAD |
|
|
rs1588538142 CA374997192 |
83 | E>D | No |
ClinGen Ensembl |
|
|
rs754446615 CA5251660 |
86 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs371317161 CA5251661 |
86 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
CA5251662 rs141709101 |
87 | T>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs754288051 CA5251696 |
91 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs749596460 CA5251697 |
92 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
COSM673978 rs751056570 CA5251699 |
94 | R>C | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed |
|
rs1261382989 CA374997361 |
98 | S>G | No |
ClinGen gnomAD |
|
|
CA374997372 rs1432801599 |
99 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA5251702 rs745709237 |
104 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5251705 rs147073375 |
108 | H>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs774630568 CA5251707 |
110 | L>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1257939333 CA374997521 |
115 | S>G | No |
ClinGen gnomAD |
|
|
CA5251708 rs761776626 |
116 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs772377103 CA5251709 |
123 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374997605 rs1564433006 |
124 | S>P | No |
ClinGen Ensembl |
|
|
rs1472739092 CA374997625 |
126 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
rs199885166 CA5251713 |
128 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs531446315 CA200314955 |
131 | Q>* | No |
ClinGen Ensembl |
|
|
rs776976149 CA5251716 |
132 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs562638413 CA5251718 |
136 | G>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs755939591 CA5251723 |
140 | I>V | No |
ClinGen ExAC gnomAD |
|
| rs1412326548 | 145 | I>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5251749 rs771187222 |
147 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs746257072 CA5251751 |
149 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1318493395 CA374997809 |
150 | M>R | No |
ClinGen TOPMed |
|
|
rs1404594512 CA374997813 |
151 | K>Q | No |
ClinGen gnomAD |
|
|
CA374997827 rs1383542677 |
153 | A>T | No |
ClinGen gnomAD |
|
|
rs1239547944 CA374997841 |
155 | V>M | No |
ClinGen gnomAD |
|
|
rs1158106390 CA374997849 |
156 | L>F | No |
ClinGen TOPMed |
|
|
rs763433765 CA5251754 |
157 | I>N | No |
ClinGen ExAC gnomAD |
|
|
rs1588538597 CA374997860 |
158 | T>A | No |
ClinGen Ensembl |
|
|
rs1356089042 CA374997863 |
158 | T>S | No |
ClinGen gnomAD |
|
|
rs1419886224 CA374997866 |
159 | R>C | No |
ClinGen TOPMed |
|
|
CA5251755 rs769188538 |
159 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769188538 CA374997868 |
159 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772642497 CA5251756 |
160 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760231401 CA5251757 |
161 | G>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 162 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374997896 rs1237129942 |
164 | K>T | No |
ClinGen TOPMed |
|
|
CA374997910 rs1176271577 |
166 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1381667644 CA374997920 |
168 | F>I | No |
ClinGen gnomAD |
|
|
CA200314996 rs764781911 |
172 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA5251758 rs766161082 |
172 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1197895718 CA374997971 |
176 | L>V | No |
ClinGen gnomAD |
|
| TCGA novel | 178 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 182 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5251763 rs758316825 |
182 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs777735060 CA5251764 |
185 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs950843863 CA200315003 |
187 | N>S | No |
ClinGen Ensembl |
|
|
CA374998077 rs757281821 |
192 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA5251766 rs757281821 |
192 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs148342102 CA374998081 CA5251768 |
193 | W>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5251769 rs766316927 |
195 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs865941710 CA200315008 |
195 | R>W | No |
ClinGen Ensembl |
|
|
CA374998111 rs1465554087 |
197 | P>L | No |
ClinGen gnomAD |
|
|
CA374998110 rs1465554087 |
197 | P>R | No |
ClinGen gnomAD |
|
|
CA5251773 rs774881075 |
201 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1270270686 CA374998134 |
201 | L>V | No |
ClinGen TOPMed |
|
|
rs1424183824 CA374998152 |
202 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
CA5251775 COSM1460133 rs770506343 |
202 | G>R | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs745340199 CA5251800 |
203 | E>* | No |
ClinGen ExAC gnomAD |
|
|
CA374998157 rs1362470829 |
203 | E>G | No |
ClinGen gnomAD |
|
|
CA5251801 rs373394121 |
204 | R>W | No |
ClinGen ESP ExAC gnomAD |
|
|
rs775163113 CA5251802 |
206 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1310495867 CA374998175 |
206 | Y>H | No |
ClinGen gnomAD |
|
|
rs763885651 CA5251804 |
207 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374998192 rs371033254 |
209 | P>S | No |
ClinGen gnomAD |
|
|
rs371033254 CA200315091 |
209 | P>T | No |
ClinGen gnomAD |
|
| rs755440676 | 210 | I>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755440676 | 210 | I>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1217556836 CA374998196 |
210 | I>V | No |
ClinGen gnomAD |
|
|
CA374998211 rs1262847634 |
212 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA374998235 rs1179525505 |
215 | A>G | No |
ClinGen TOPMed |
|
|
CA374998241 rs1446469808 |
216 | G>A | No |
ClinGen gnomAD |
|
|
rs1241846326 CA374998237 |
216 | G>R | No |
ClinGen gnomAD |
|
|
rs1241846326 CA374998239 |
216 | G>W | No |
ClinGen gnomAD |
|
|
rs1379816585 CA374998243 |
217 | C>G | No |
ClinGen gnomAD |
|
|
CA374998272 rs1345500468 |
220 | A>V | No |
ClinGen gnomAD |
|
|
rs1253842556 CA374998275 |
221 | E>* | No |
ClinGen TOPMed |
|
|
rs1465373889 CA374998287 |
222 | M>I | No |
ClinGen gnomAD |
|
|
CA374998303 rs1300964249 |
224 | T>S | No |
ClinGen gnomAD |
|
|
CA5251809 rs767418586 VAR_082140 |
225 | R>C | Variant assessed as Somatic; 0.0 impact. found in patients with global developmental delay and epilepsy with history of choanal atresia; unknown pathological significance [NCI-TCGA, UniProt] | No |
ClinGen UniProt ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA5251810 rs750537619 |
225 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA200315108 rs767418586 |
225 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1317836668 CA374998333 |
229 | M>T | No |
ClinGen gnomAD |
|
| VAR_013456 | 231 | G>A | No | UniProt | |
|
CA374998351 rs1261706276 |
231 | G>D | No |
ClinGen gnomAD |
|
|
CA374998346 rs1564433291 |
231 | G>S | No |
ClinGen Ensembl |
|
|
CA374998364 rs141577691 |
233 | T>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5251814 rs141577691 |
233 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1564433307 CA374998378 |
235 | Q>R | No |
ClinGen Ensembl |
|
|
CA5251817 rs372825047 |
237 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA374998389 rs372825047 |
237 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5251820 rs11554104 |
239 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs11554104 CA200315135 |
239 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA374998413 rs1431221428 |
241 | I>V | No |
ClinGen gnomAD |
|
|
CA5251822 rs775012342 |
242 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA374998428 rs1397413665 |
243 | Q>* | No |
ClinGen gnomAD |
|
| TCGA novel | 245 | C>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768177225 CA374998448 |
246 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768177225 CA5251824 |
246 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374998459 rs1588538886 |
248 | I>L | No |
ClinGen Ensembl |
|
|
rs774177908 CA5251825 |
249 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA374998471 rs761561096 |
250 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1257998616 CA374998474 |
250 | P>R | No |
ClinGen gnomAD |
|
|
rs761561096 CA5251826 |
250 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374998472 rs761561096 |
250 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770932885 CA5251867 |
252 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs1156517816 CA374998507 |
253 | W>* | No |
ClinGen gnomAD |
|
|
rs776385767 CA5251868 |
254 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA374998524 rs769930999 |
256 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs769930999 CA5251870 |
256 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA374998532 rs1588539183 |
257 | D>G | No |
ClinGen Ensembl |
|
|
rs1406035499 CA374998528 |
257 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs775710179 CA5251871 |
258 | N>H | No |
ClinGen ExAC gnomAD |
|
|
rs763329642 CA374998539 |
258 | N>I | No |
ClinGen ExAC gnomAD |
|
|
rs763329642 CA5251872 |
258 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs775710179 CA374998537 |
258 | N>Y | No |
ClinGen ExAC gnomAD |
|
|
rs751991570 CA5251875 |
263 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs751991570 CA5251874 |
263 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA374998586 rs1436815577 |
265 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA200315474 rs763851751 |
267 | L>P | No |
ClinGen Ensembl |
|
|
rs768088819 CA5251876 |
268 | V>A | No |
ClinGen ExAC |
|
|
rs768690569 CA200315479 |
270 | G>C | No |
ClinGen Ensembl |
|
|
CA374998621 rs1564433574 |
271 | Q>* | No |
ClinGen Ensembl |
|
| TCGA novel | 271 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1243217909 CA374998637 |
273 | R>Q | No |
ClinGen gnomAD |
|
|
COSM1105407 rs376244572 CA5251878 |
273 | R>W | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA gnomAD |
|
rs1588539233 CA374998653 |
275 | V>G | No |
ClinGen Ensembl |
|
|
CA374998690 rs1365208357 |
281 | A>T | No |
ClinGen gnomAD |
|
|
CA5251882 rs758087169 |
283 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370938433 COSM1460136 CA200315487 |
284 | R>C | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ESP NCI-TCGA |
|
CA374998727 rs1564433598 |
286 | P>L | No |
ClinGen Ensembl |
|
|
rs1206439670 CA590939019 |
287 | Y>* | No |
ClinGen gnomAD |
|
|
rs781196606 CA374998737 |
288 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781196606 CA5251886 |
288 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374998740 rs1218758923 |
288 | A>V | No |
ClinGen TOPMed |
|
|
rs745870791 CA5251887 |
289 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs775765477 CA5251889 |
293 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs775765477 CA5251890 |
293 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs774385639 CA5251892 |
294 | K>M | No |
ClinGen ExAC gnomAD |
|
|
CA5251894 rs145977259 |
298 | L>P | No |
ClinGen ESP ExAC gnomAD |
|
|
CA374998802 rs1588539291 |
299 | D>A | No |
ClinGen Ensembl |
|
|
CA374998806 rs1230213286 |
299 | D>E | No |
ClinGen TOPMed |
|
|
rs773619937 CA5251895 |
300 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs761228208 CA5251896 |
302 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs139856271 CA5251898 |
304 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5251897 rs766848600 |
304 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs763822642 CA5251900 |
306 | S>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA374998918 rs1430649434 |
309 | A>P | No |
ClinGen gnomAD |
|
| TCGA novel | 309 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1461954326 CA374998929 |
310 | L>F | No |
ClinGen gnomAD |
|
|
CA374998979 rs1564433653 |
314 | F>V | No |
ClinGen Ensembl |
|
|
rs1564433655 CA374998983 |
314 | F>Y | No |
ClinGen Ensembl |
|
|
CA200315499 rs868437001 |
318 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs1362073818 CA374999044 |
319 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1302788481 CA374999066 |
320 | M>I | No |
ClinGen gnomAD |
|
|
rs756164503 CA5251905 |
321 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA5251907 rs749397034 |
322 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA5251909 rs774633888 COSM48296 |
323 | D>N | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA374999113 rs1588539341 |
325 | K>E | No |
ClinGen Ensembl |
|
|
rs1339484627 CA374999119 |
325 | K>R | No |
ClinGen gnomAD |
|
|
CA374999129 rs1564433682 |
326 | G>D | No |
ClinGen Ensembl |
|
|
CA200315511 rs1004550576 |
327 | M>I | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 328 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374999156 rs1289167649 |
328 | L>P | No |
ClinGen gnomAD |
|
|
rs1588539353 CA374999168 |
330 | T>P | No |
ClinGen Ensembl |
|
|
rs1451159017 CA374999180 |
331 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA374999203 rs1489949745 |
333 | T>M | No |
ClinGen gnomAD |
|
|
CA374999238 rs1428423917 |
336 | F>S | No |
ClinGen gnomAD |
|
|
rs760988808 CA5251913 |
337 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA374999311 rs1363983638 |
342 | P>L | No |
ClinGen gnomAD |
|
| TCGA novel | 344 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1426836717 CA374999328 |
344 | R>Q | No |
ClinGen gnomAD |
|
|
rs1218615279 CA374999351 |
346 | G>A | No |
ClinGen TOPMed |
|
|
CA374999350 rs1218615279 |
346 | G>D | No |
ClinGen TOPMed |
|
|
CA5251916 rs762456826 |
346 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs763581624 CA5251917 |
347 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA5251919 rs756913144 |
349 | I>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 352 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5251920 rs200376810 |
354 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374999441 rs200376810 |
354 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755920229 CA5251922 |
355 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374999480 rs1222840513 |
357 | S>T | No |
ClinGen gnomAD |
|
|
rs780150586 CA5251923 |
358 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753758776 CA5251924 |
358 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs754937212 CA5251925 |
359 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs778916606 CA5251927 |
360 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA5251928 rs748237241 |
360 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs777924310 CA5251930 |
361 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs747387277 CA5251931 |
362 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1362949448 CA374999533 |
362 | T>I | No |
ClinGen gnomAD |
|
|
CA374999526 rs747387277 |
362 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1451386730 CA374999538 |
363 | T>A | No |
ClinGen gnomAD |
|
|
CA374999540 rs771220295 |
363 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA5251932 rs771220295 |
363 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs776977894 CA5251933 |
366 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5251935 rs201475786 |
368 | F>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA374999623 rs1384441244 |
370 | R>C | No |
ClinGen gnomAD |
|
|
rs1246168747 CA374999624 |
370 | R>H | Variant assessed as Somatic; 4.67e-05 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA5251937 rs761350967 |
371 | V>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 373 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with P50750
5 regional properties for P50750
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Helix-turn-helix motif | 124 - 149 | IPR000047 |
| domain | Homeobox domain | 93 - 157 | IPR001356 |
| conserved_site | Homeobox, conserved site | 128 - 151 | IPR017970 |
| domain | Homeobox domain, metazoa | 117 - 128 | IPR020479-1 |
| domain | Homeobox domain, metazoa | 132 - 151 | IPR020479-2 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.22 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cyclin/CDK positive transcription elongation factor complex | A transcription elongation factor complex that facilitates the transition from abortive to productive elongation by phosphorylating the CTD domain of the large subunit of DNA-directed RNA polymerase II, holoenzyme. Contains a cyclin and a cyclin-dependent protein kinase catalytic subunit. |
| cytoplasmic ribonucleoprotein granule | A ribonucleoprotein granule located in the cytoplasm. |
| mediator complex | A protein complex that interacts with the carboxy-terminal domain of the largest subunit of RNA polymerase II and plays an active role in transducing the signal from a transcription factor to the transcriptional machinery. The mediator complex is required for activation of transcription of most protein-coding genes, but can also act as a transcriptional corepressor. The Saccharomyces complex contains several identifiable subcomplexes: a head domain comprising Srb2, -4, and -5, Med6, -8, and -11, and Rox3 proteins; a middle domain comprising Med1, -4, and -7, Nut1 and -2, Cse2, Rgr1, Soh1, and Srb7 proteins; a tail consisting of Gal11p, Med2p, Pgd1p, and Sin4p; and a regulatory subcomplex comprising Ssn2, -3, and -8, and Srb8 proteins. Metazoan mediator complexes have similar modular structures and include homologs of yeast Srb and Med proteins. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| P-TEFb complex | A dimeric positive transcription elongation factor complex b that comprises a cyclin-dependent kinase containing the catalytic subunit, Cdk9, and a regulatory subunit, cyclin T. |
| PML body | A class of nuclear body; they react against SP100 auto-antibodies (PML, promyelocytic leukemia); cells typically contain 10-30 PML bodies per nucleus; alterations in the localization of PML bodies occurs after viral infection. |
| transcription elongation factor complex | Any protein complex that interacts with RNA polymerase II to increase (positive transcription elongation factor) or reduce (negative transcription elongation factor) the rate of transcription elongation. |
12 GO annotations of molecular function
| Name | Definition |
|---|---|
| 7SK snRNA binding | Binding to a 7SK small nuclear RNA (7SK snRNA). |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| cyclin-dependent protein serine/threonine kinase activity | Cyclin-dependent catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II CTD heptapeptide repeat kinase activity | Catalysis of the reaction: ATP + RNA polymerase II large subunit CTD heptapeptide repeat (YSPTSPS) = ADP + H+ + phosphorylated RNA polymerase II. |
| transcription coactivator binding | Binding to a transcription coactivator, a protein involved in positive regulation of transcription via protein-protein interactions with transcription factors and other proteins that positively regulate transcription. Transcription coactivators do not bind DNA directly, but rather mediate protein-protein interactions between activating transcription factors and the basal transcription machinery. |
22 GO annotations of biological process
| Name | Definition |
|---|---|
| cell population proliferation | The multiplication or reproduction of cells, resulting in the expansion of a cell population. |
| cellular response to cytokine stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| DNA repair | The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway. |
| phosphorylation of RNA polymerase II C-terminal domain serine 2 residues involved in positive regulation of transcription elongation from RNA polymerase II promoter | Any phosphorylation of RNA polymerase II C-terminal domain serine 2 residues that is involved in positive regulation of transcription elongation from RNA polymerase II promoter. |
| phosphorylation of RNA polymerase II C-terminal domain serine 5 residues involved in positive regulation of transcription elongation from RNA polymerase II promoter | Any phosphorylation of RNA polymerase II C-terminal domain serine 5 residues that is involved in positive regulation of transcription elongation from RNA polymerase II promoter. |
| positive regulation by host of viral transcription | Any process in which a host organism activates or increases the frequency, rate or extent of viral transcription, the synthesis of either RNA on a template of DNA or DNA on a template of RNA. |
| positive regulation of cardiac muscle hypertrophy | Any process that increases the rate, frequency or extent of the enlargement or overgrowth of all or part of the heart due to an increase in size (not length) of individual cardiac muscle fibers, without cell division. |
| positive regulation of mRNA 3'-UTR binding | Any process that activates or increases the frequency, rate or extent of mRNA 3'-UTR binding. |
| positive regulation of protein localization to chromatin | Any process that activates or increases the frequency, rate or extent of protein localization to chromatin. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription elongation by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription elongation, the extension of an RNA molecule after transcription initiation and promoter clearance by the addition of ribonucleotides, catalyzed by RNA polymerase II. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of DNA repair | Any process that modulates the frequency, rate or extent of DNA repair. |
| regulation of histone modification | Any process that modulates the frequency, rate or extent of the covalent alteration of a histone. |
| regulation of mRNA 3'-end processing | Any process that modulates the frequency, rate or extent of mRNA 3'-end processing, any process involved in forming the mature 3' end of an mRNA molecule. |
| regulation of muscle cell differentiation | Any process that modulates the frequency, rate or extent of muscle cell differentiation. |
| replication fork processing | The process in which a DNA replication fork that has stalled is restored to a functional state and replication is restarted. The stalling may be due to DNA damage, DNA secondary structure, bound proteins, dNTP shortage, or other causes. |
| response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| transcription by RNA polymerase II | The synthesis of RNA from a DNA template by RNA polymerase II (RNAP II), originating at an RNA polymerase II promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs). |
| transcription elongation by RNA polymerase II promoter | The extension of an RNA molecule after transcription initiation and promoter clearance at an RNA polymerase II promoter by the addition of ribonucleotides catalyzed by RNA polymerase II. |
| transcription initiation at RNA polymerase II promoter | A transcription initiation process that takes place at a RNA polymerase II gene promoter. Messenger RNAs (mRNA) genes, as well as some non-coding RNAs, are transcribed by RNA polymerase II. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5EAB2 | CDK9 | Cyclin-dependent kinase 9 | Bos taurus (Bovine) | PR |
| Q5ZKN1 | CDK9 | Cyclin-dependent kinase 9 | Gallus gallus (Chicken) | PR |
| Q15131 | CDK10 | Cyclin-dependent kinase 10 | Homo sapiens (Human) | PR |
| Q9UQ88 | CDK11A | Cyclin-dependent kinase 11A | Homo sapiens (Human) | PR |
| Q00526 | CDK3 | Cyclin-dependent kinase 3 | Homo sapiens (Human) | PR |
| Q96Q40 | CDK15 | Cyclin-dependent kinase 15 | Homo sapiens (Human) | PR |
| O94921 | CDK14 | Cyclin-dependent kinase 14 | Homo sapiens (Human) | PR |
| Q00537 | CDK17 | Cyclin-dependent kinase 17 | Homo sapiens (Human) | PR |
| P49336 | CDK8 | Cyclin-dependent kinase 8 | Homo sapiens (Human) | PR |
| Q9BWU1 | CDK19 | Cyclin-dependent kinase 19 | Homo sapiens (Human) | PR |
| Q5MAI5 | CDKL4 | Cyclin-dependent kinase-like 4 | Homo sapiens (Human) | PR |
| Q00532 | CDKL1 | Cyclin-dependent kinase-like 1 | Homo sapiens (Human) | PR |
| Q92772 | CDKL2 | Cyclin-dependent kinase-like 2 | Homo sapiens (Human) | PR |
| Q8IZL9 | CDK20 | Cyclin-dependent kinase 20 | Homo sapiens (Human) | PR |
| P21127 | CDK11B | Cyclin-dependent kinase 11B | Homo sapiens (Human) | PR |
| Q99J95 | Cdk9 | Cyclin-dependent kinase 9 | Mus musculus (Mouse) | PR |
| Q641Z4 | Cdk9 | Cyclin-dependent kinase 9 | Rattus norvegicus (Rat) | PR |
| F4I114 | At1g09600 | Probable serine/threonine-protein kinase At1g09600 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZVM9 | At1g54610 | Probable serine/threonine-protein kinase At1g54610 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6GLD8 | cdk9 | Cyclin-dependent kinase 9 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAKQYDSVEC | PFCDEVSKYE | KLAKIGQGTF | GEVFKARHRK | TGQKVALKKV | LMENEKEGFP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ITALREIKIL | QLLKHENVVN | LIEICRTKAS | PYNRCKGSIY | LVFDFCEHDL | AGLLSNVLVK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FTLSEIKRVM | QMLLNGLYYI | HRNKILHRDM | KAANVLITRD | GVLKLADFGL | ARAFSLAKNS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QPNRYTNRVV | TLWYRPPELL | LGERDYGPPI | DLWGAGCIMA | EMWTRSPIMQ | GNTEQHQLAL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ISQLCGSITP | EVWPNVDNYE | LYEKLELVKG | QKRKVKDRLK | AYVRDPYALD | LIDKLLVLDP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AQRIDSDDAL | NHDFFWSDPM | PSDLKGMLST | HLTSMFEYLA | PPRRKGSQIT | QQSTNQSRNP |
| 370 | |||||
| ATTNQTEFER | VF |