P50148
Gene name |
GNAQ (GAQ) |
Protein name |
Guanine nucleotide-binding protein G |
Names |
q subunit alpha , Guanine nucleotide-binding protein alpha-q |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2776 |
EC number |
|
Protein Class |
|
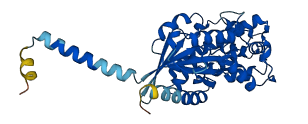
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
189-353 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Goricanec D et al. (2016) "Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding", Proceedings of the National Academy of Sciences of the United States of America, 113, E3629-38
- Coleman DE et al. (1999) "Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex", The Journal of biological chemistry, 274, 16669-72
- Lutz S et al. (2007) "Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs", Science (New York, N.Y.), 318, 1923-7
Autoinhibited structure

Activated structure

20 structures for P50148
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6VU5 | EM | 350 A | B | 1-359 | PDB |
| 7EZM | EM | 290 A | A | 37-359 | PDB |
| 7F6G | EM | 290 A | B | 2-359 | PDB |
| 7F6H | EM | 290 A | B | 2-359 | PDB |
| 7F6I | EM | 280 A | B | 2-359 | PDB |
| 7F8W | EM | 310 A | A | 29-359 | PDB |
| 7W3Z | EM | 300 A | B | 36-359 | PDB |
| 7W40 | EM | 300 A | B | 36-359 | PDB |
| 7XOW | EM | 310 A | A | 36-359 | PDB |
| 8G59 | EM | 264 A | A | 335-359 | PDB |
| 8IA7 | EM | 310 A | A | 36-359 | PDB |
| 8IYS | EM | 295 A | A | 36-359 | PDB |
| 8J9N | EM | 350 A | C | 345-359 | PDB |
| 8JPB | EM | 307 A | Q | 37-359 | PDB |
| 8JPC | EM | 307 A | Q | 37-359 | PDB |
| 8JPE | EM | 291 A | Q | 37-359 | PDB |
| 8SZG | EM | 360 A | C | 29-359 | PDB |
| 8UQN | EM | 340 A | A | 7-359 | PDB |
| 8UQO | EM | 337 A | A | 7-359 | PDB |
| AF-P50148-F1 | Predicted | AlphaFoldDB |
143 variants for P50148
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV002254275 RCV001526543 RCV000043592 VAR_067270 RCV003221795 COSM52975 CA143805 RCV000533476 rs397514698 RCV001526638 RCV000043593 RCV002294003 RCV001705695 |
183 | R>Q | eye Angioosteohypertrophic syndrome Capillary malformation Familial multiple nevi flammei large_intestine Melanoma Variant assessed as Somatic; impact. Sturge-Weber syndrome meninges SWS; found as somatic mosaic mutation in CMC; also found in melanocytomas sample; somatic mutation; shows significant activation of EPHB2 compared to control [Cosmic, ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl NCI-TCGA dbSNP |
|
RCV000441114 rs1057519853 CA16602757 COSM28757 |
209 | Q>L | eye skin Uveal melanoma meninges [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
RCV000436244 VAR_067271 RCV000426416 COSM28757 CA16602434 rs121913492 |
209 | Q>L | eye Melanoma Variant assessed as Somatic; impact. skin Uveal melanoma meninges found in blue naevi and uveal melanoma samples; somatic mutation; constitutive activation [Cosmic, ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl NCI-TCGA dbSNP |
|
COSM28758 CA16602436 rs121913492 RCV000436033 RCV000425312 |
209 | Q>P | eye NS Melanoma Variant assessed as Somatic; impact. central_nervous_system skin Uveal melanoma meninges [Cosmic, ClinVar, NCI-TCGA] | Yes |
ClinGen cosmic curated ClinVar Ensembl NCI-TCGA dbSNP |
|
RCV001327979 CA16602435 COSM28760 rs121913492 RCV000442822 |
209 | Q>R | eye Melanoma [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
rs780163661 CA5094751 |
2 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA194745058 rs1059518 |
4 | E>D | No |
ClinGen Ensembl |
|
|
rs746420387 CA373999009 |
6 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA5094749 rs746420387 |
6 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA373999005 rs779515663 |
6 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1264916735 CA373999008 |
6 | I>T | No |
ClinGen TOPMed |
|
|
rs746420387 CA5094750 |
6 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA5094747 rs757711264 |
7 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1203392784 CA373998996 |
8 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 10 | C>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 35 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5094738 rs762737987 |
42 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1466271329 CA373998754 |
43 | L>P | No |
ClinGen gnomAD |
|
| TCGA novel | 48 | G>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 50 | S>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs773781296 CA5094692 |
59 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA373997074 rs1256109740 |
69 | D>E | No |
ClinGen gnomAD |
|
|
CA5094688 rs768341234 |
76 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA5094687 rs746699009 |
77 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA373997010 rs1369065497 |
79 | V>L | No |
ClinGen gnomAD |
|
|
rs1251821572 CA373997000 |
80 | Y>F | No |
ClinGen TOPMed |
|
|
CA373996971 rs1192458704 |
84 | F>Y | No |
ClinGen TOPMed |
|
|
CA5094686 COSM1463120 rs761634659 |
85 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA5094683 rs778978521 |
87 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373996946 rs1191803927 |
88 | Q>E | No |
ClinGen Ensembl |
|
|
rs1136722 CA194732163 |
92 | R>T | No |
ClinGen Ensembl |
|
|
rs1444819549 CA373996911 |
93 | A>T | No |
ClinGen gnomAD |
|
|
CA5094682 rs757129673 |
95 | D>E | No |
ClinGen ExAC gnomAD |
|
|
COSM404628 CA5094681 rs753716491 |
96 | T>S | lung Variant assessed as Somatic; 0.004093 impact. prostate [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA373996847 rs1416428102 |
99 | I>M | No |
ClinGen gnomAD |
|
|
CA5094677 rs766649624 |
100 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA5094678 rs766649624 |
100 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA5094675 rs200106152 |
101 | Y>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA373996777 COSM3664424 rs1468666506 COSM3664425 |
102 | K>N | liver [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA5094674 rs765827868 |
103 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA194732161 rs77227613 |
103 | Y>S | No |
ClinGen Ensembl |
|
|
CA194732160 rs200658460 |
104 | E>G | No |
ClinGen 1000Genomes |
|
|
rs1438658973 CA373996734 |
105 | H>D | No |
ClinGen TOPMed |
|
|
CA5094673 rs762200216 |
107 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA5094652 rs377726080 |
108 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1288502233 CA373998605 |
109 | H>N | No |
ClinGen gnomAD |
|
|
CA5094651 rs761095319 |
109 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775821091 CA5094650 |
110 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA373998579 rs1430429958 |
113 | V>I | No |
ClinGen TOPMed |
|
|
CA5094648 rs759080789 |
114 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA194719766 rs1010141343 |
114 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1425322060 CA373998570 |
115 | E>K | No |
ClinGen gnomAD |
|
| TCGA novel | 116 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373998561 rs1383379438 |
116 | V>L | No |
ClinGen TOPMed |
|
|
CA5094647 rs371301832 |
117 | D>G | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1351194942 CA373998517 |
122 | S>C | No |
ClinGen TOPMed |
|
|
rs1163999044 CA373998520 |
122 | S>P | No |
ClinGen TOPMed |
|
|
CA5094643 RCV000912112 rs192927818 |
130 | D>N | No |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs1362009464 CA373998455 |
131 | A>G | No |
ClinGen TOPMed |
|
|
CA5094642 rs748011987 |
134 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA373998429 rs1305477251 COSM281412 |
135 | L>V | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 137 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1295375779 CA373998363 |
144 | C>R | No |
ClinGen gnomAD |
|
| TCGA novel | 150 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5094638 rs779143139 |
151 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs200924131 CA5094636 |
152 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA194719764 rs923027293 |
157 | T>A | No |
ClinGen Ensembl |
|
| TCGA novel | 159 | Y>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1215304254 CA373998258 |
159 | Y>H | No |
ClinGen TOPMed |
|
| TCGA novel | 161 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373998189 rs1587920943 |
165 | D>A | No |
ClinGen Ensembl |
|
|
rs1402276757 COSM1253350 CA373998179 |
167 | V>I | Variant assessed as Somatic; 0.0 impact. oesophagus [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs777801390 CA5094618 |
168 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA373998165 rs1587920931 |
169 | D>A | No |
ClinGen Ensembl |
|
|
rs756147804 CA5094617 |
170 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA194717624 rs980935611 |
170 | P>R | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 171 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373998146 rs1393447827 |
172 | Y>C | No |
ClinGen TOPMed |
|
|
rs1587920918 CA373998130 |
175 | T>A | No |
ClinGen Ensembl |
|
|
CA373998128 rs1461816732 |
175 | T>K | No |
ClinGen TOPMed |
|
|
CA194717622 rs28764015 |
191 | E>K | No |
ClinGen Ensembl |
|
| TCGA novel | 192 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 197 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373997925 rs1223047296 |
203 | M>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA373997905 rs1476186591 |
206 | V>I | No |
ClinGen TOPMed |
|
|
CA373997787 rs1423930996 |
222 | N>S | No |
ClinGen TOPMed |
|
|
rs769002669 CA194717256 |
226 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
| TCGA novel | 235 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373997689 rs1299114921 |
236 | D>E | No |
ClinGen TOPMed |
|
|
rs1587919416 CA373997675 |
238 | V>A | No |
ClinGen Ensembl |
|
|
rs968608243 CA373997666 |
240 | V>L | No |
ClinGen TOPMed |
|
|
rs968608243 CA194717254 |
240 | V>M | No |
ClinGen TOPMed |
|
|
rs1361664931 CA373997649 |
242 | S>L | No |
ClinGen gnomAD |
|
|
CA5094584 rs759267731 |
244 | N>S | No |
ClinGen ExAC TOPMed |
|
| rs1449750349 | 246 | N>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 248 | M>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1587886275 CA373997588 |
249 | E>G | No |
ClinGen Ensembl |
|
| TCGA novel | 250 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA194710022 rs2013373 |
256 | R>K | No |
ClinGen gnomAD |
|
|
CA373997537 rs2013373 |
256 | R>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs149046601 CA5094557 |
259 | I>V | No |
ClinGen ESP ExAC TOPMed |
|
|
CA373997495 rs1285850636 |
262 | P>L | No |
ClinGen TOPMed |
|
|
rs200517432 CA194710020 |
264 | F>L | No |
ClinGen 1000Genomes |
|
|
rs372073168 CA5094556 |
265 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs765491785 CA5094555 |
267 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA194710019 rs1059522 |
272 | F>L | No |
ClinGen Ensembl |
|
|
rs898844416 CA194710018 |
277 | D>Y | No |
ClinGen TOPMed |
|
|
CA194710017 rs1059523 |
279 | L>P | No |
ClinGen Ensembl |
|
| TCGA novel | 279 | L>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373997378 rs1182621194 |
280 | E>Q | No |
ClinGen TOPMed |
|
|
CA5094553 rs776673486 |
284 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA373997348 rs1273681754 |
284 | M>V | No |
ClinGen gnomAD |
|
|
rs886255565 CA194710016 |
288 | L>I | No |
ClinGen TOPMed gnomAD |
|
|
CA194710015 rs201195349 |
292 | F>S | No |
ClinGen 1000Genomes |
|
|
CA5094533 rs374303316 |
299 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373996515 rs1311795673 |
300 | R>G | No |
ClinGen TOPMed |
|
|
rs760869735 CA5094532 |
300 | R>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1236524878 CA373996508 |
301 | D>N | No |
ClinGen gnomAD |
|
|
rs772749242 CA5094530 |
302 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs774951302 CA5094528 |
303 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs777546823 CA5094526 |
304 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA5094527 rs376987002 |
304 | A>P | No |
ClinGen ESP ExAC gnomAD |
|
|
rs777546823 CA5094525 |
304 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA5094524 rs769315863 |
306 | R>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 307 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 307 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1246606508 CA373996442 CA373996441 |
311 | K>N | No |
ClinGen TOPMed |
|
|
rs372746608 CA5094522 |
312 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA5094520 rs751516475 |
317 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs183713546 CA5094519 |
319 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1352440197 CA373996392 |
319 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1216739414 CA373996370 |
321 | D>E | No |
ClinGen TOPMed |
|
|
rs397825484 CA194709238 |
322 | K>Q | No |
ClinGen Ensembl |
|
|
rs1297612239 CA373996349 |
324 | I>M | No |
ClinGen gnomAD |
|
| TCGA novel | 326 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373996304 rs1238540285 |
331 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 333 | D>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1412853649 CA373996278 |
335 | E>K | No |
ClinGen gnomAD |
|
|
CA194709237 rs1059527 |
337 | I>V | No |
ClinGen Ensembl |
|
|
rs1180028656 CA373996254 |
338 | R>C | No |
ClinGen TOPMed |
|
|
CA373996215 rs1257829666 |
344 | V>I | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 347 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA194709234 rs1059531 VAR_059319 |
355 | E>D | No |
ClinGen UniProt Ensembl dbSNP |
2 associated diseases with P50148
[MIM: 163000]: Capillary malformations, congenital (CMC)
A form of vascular malformations that are present from birth, tend to grow with the individual, do not regress spontaneously, and show normal rates of endothelial cell turnover. Capillary malformations are distinct from capillary hemangiomas, which are highly proliferative lesions that appear shortly after birth and show rapid growth, slow involution, and endothelial hypercellularity. . Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 185300]: Sturge-Weber syndrome (SWS)
A syndrome characterized by an intracranial vascular anomaly, leptomeningeal angiomatosis, most often involving the occipital and posterior parietal lobes. The most common features are facial cutaneous vascular malformations (port-wine stains), seizures, and glaucoma. Stasis results in ischemia underlying the leptomeningeal angiomatosis, leading to calcification and laminar cortical necrosis. The clinical course is highly variable and some children experience intractable seizures, intellectual disability, and recurrent stroke-like episodes. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of vascular malformations that are present from birth, tend to grow with the individual, do not regress spontaneously, and show normal rates of endothelial cell turnover. Capillary malformations are distinct from capillary hemangiomas, which are highly proliferative lesions that appear shortly after birth and show rapid growth, slow involution, and endothelial hypercellularity. . Note=The disease is caused by variants affecting the gene represented in this entry.
- A syndrome characterized by an intracranial vascular anomaly, leptomeningeal angiomatosis, most often involving the occipital and posterior parietal lobes. The most common features are facial cutaneous vascular malformations (port-wine stains), seizures, and glaucoma. Stasis results in ischemia underlying the leptomeningeal angiomatosis, leading to calcification and laminar cortical necrosis. The clinical course is highly variable and some children experience intractable seizures, intellectual disability, and recurrent stroke-like episodes. . Note=The disease is caused by variants affecting the gene represented in this entry.
No regional properties for P50148
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P50148 | |||
Functions
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| nuclear membrane | Either of the lipid bilayers that surround the nucleus and form the nuclear envelope; excludes the intermembrane space. |
| photoreceptor outer segment | The outer segment of a vertebrate photoreceptor that contains a stack of membrane discs embedded with photoreceptor proteins. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activator activity | Binds to and increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP. |
| GTPase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| action potential | A process in which membrane potential cycles through a depolarizing spike, triggered in response to depolarization above some threshold, followed by repolarization. This cycle is driven by the flow of ions through various voltage gated channels with different thresholds and ion specificities. |
| activation of phospholipase C activity | The initiation of the activity of the inactive enzyme phospolipase C as the result of The series of molecular signals generated as a consequence of a G protein-coupled receptor binding to its physiological ligand. |
| adenylate cyclase-activating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation of adenylyl cyclase activity and a subsequent increase in the intracellular concentration of cyclic AMP (cAMP). |
| blood coagulation | The sequential process in which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages |
| entrainment of circadian clock | The synchronization of a circadian rhythm to environmental time cues such as light. |
| G protein-coupled acetylcholine receptor signaling pathway | A G protein-coupled receptor signaling pathway initiated by a ligand binding to an acetylcholine receptor on the surface of a target cell, and ends with regulation of a downstream cellular process, e.g. transcription. |
| glutamate receptor signaling pathway | The series of molecular signals initiated by the binding of glutamate to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| negative regulation of protein kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase activity. |
| phospholipase C-activating dopamine receptor signaling pathway | A phospholipase C-activating receptor G protein-coupled receptor signaling pathway initiated by dopamine binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| phototransduction, visible light | The sequence of reactions within a cell required to convert absorbed photons from visible light into a molecular signal. A visible light stimulus is electromagnetic radiation that can be perceived visually by an organism; for organisms lacking a visual system, this can be defined as light with a wavelength within the range 380 to 780 nm. |
| protein stabilization | Any process involved in maintaining the structure and integrity of a protein and preventing it from degradation or aggregation. |
| regulation of canonical Wnt signaling pathway | Any process that modulates the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| regulation of platelet activation | Any process that modulates the rate or frequency of platelet activation. Platelet activation is a series of progressive, overlapping events triggered by exposure of the platelets to subendothelial tissue. |
30 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38408 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Bos taurus (Bovine) | PR |
| Q28294 | GNAQ | Guanine nucleotide-binding protein G(q) subunit alpha | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P23625 | Galphaq | G protein alpha q subunit | Drosophila melanogaster (Fruit fly) | PR |
| O95837 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Homo sapiens (Human) | SS |
| P29992 | GNA11 | Guanine nucleotide-binding protein subunit alpha-11 | Homo sapiens (Human) | SS |
| P30679 | GNA15 | Guanine nucleotide-binding protein subunit alpha-15 | Homo sapiens (Human) | SS |
| P11488 | GNAT1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| P38405 | GNAL | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P63096 | GNAI1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | EV |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P30678 | Gna15 | Guanine nucleotide-binding protein subunit alpha-15 | Mus musculus (Mouse) | PR |
| P21279 | Gnaq | Guanine nucleotide-binding protein G(q) subunit alpha | Mus musculus (Mouse) | PR |
| Q2PKF4 | GNAQ | Guanine nucleotide-binding protein G(q) subunit alpha | Sus scrofa (Pig) | PR |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| O88302 | Gna15 | Guanine nucleotide-binding protein subunit alpha-15 | Rattus norvegicus (Rat) | SS |
| Q9JID2 | Gna11 | Guanine nucleotide-binding protein subunit alpha-11 | Rattus norvegicus (Rat) | SS |
| P82471 | Gnaq | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTLESIMACC | LSEEAKEARR | INDEIERQLR | RDKRDARREL | KLLLLGTGES | GKSTFIKQMR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IIHGSGYSDE | DKRGFTKLVY | QNIFTAMQAM | IRAMDTLKIP | YKYEHNKAHA | QLVREVDVEK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VSAFENPYVD | AIKSLWNDPG | IQECYDRRRE | YQLSDSTKYY | LNDLDRVADP | AYLPTQQDVL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RVRVPTTGII | EYPFDLQSVI | FRMVDVGGQR | SERRKWIHCF | ENVTSIMFLV | ALSEYDQVLV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESDNENRMEE | SKALFRTIIT | YPWFQNSSVI | LFLNKKDLLE | EKIMYSHLVD | YFPEYDGPQR |
| 310 | 320 | 330 | 340 | 350 | |
| DAQAAREFIL | KMFVDLNPDS | DKIIYSHFTC | ATDTENIRFV | FAAVKDTILQ | LNLKEYNLV |