P49703
Gene name |
ARL4D (ARF4L) |
Protein name |
ADP-ribosylation factor-like protein 4D |
Names |
ADP-ribosylation factor-like protein 4L |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:379 |
EC number |
|
Protein Class |
|
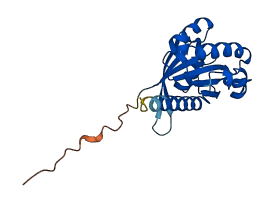
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P49703
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P49703-F1 | Predicted | AlphaFoldDB |
132 variants for P49703
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA399902082 rs1333702358 |
4 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA399902090 rs1343784186 |
5 | L>W | No |
ClinGen TOPMed gnomAD |
|
|
CA8591062 rs763660320 |
6 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs376543478 CA8591063 |
8 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs756872247 CA8591065 |
10 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8591064 rs756872247 |
10 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs150448633 CA8591067 |
12 | A>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370249858 CA8591066 |
12 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs150448633 CA399902135 |
12 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8591069 rs748735507 |
13 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772216888 CA8591070 |
15 | F>C | No |
ClinGen ExAC gnomAD |
|
|
rs773181489 CA8591071 |
15 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1339283176 CA399902162 |
17 | P>S | No |
ClinGen gnomAD |
|
|
CA399902166 rs1455113750 |
18 | H>N | No |
ClinGen TOPMed |
|
|
rs747203431 CA8591072 |
18 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs367828140 CA8591073 |
21 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs776948814 CA8591074 |
23 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs759300772 CA8591075 |
24 | V>I | No |
ClinGen ExAC |
|
|
rs200323250 CA290575609 |
27 | I>T | No |
ClinGen TOPMed |
|
|
CA290575613 rs368264928 |
31 | S>C | No |
ClinGen ESP TOPMed |
|
|
rs1598080146 CA399902273 |
35 | T>P | No |
ClinGen Ensembl |
|
|
rs1567909605 CA399902281 |
36 | S>T | No |
ClinGen Ensembl |
|
|
CA399902287 rs1359999866 |
37 | L>F | No |
ClinGen gnomAD |
|
|
rs1373132732 CA399902292 |
38 | L>V | No |
ClinGen TOPMed |
|
|
rs1300929529 CA399902306 |
40 | R>G | No |
ClinGen TOPMed |
|
|
rs767054676 CA8591082 |
40 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA8591083 rs750148648 |
41 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs371969086 CA290575619 |
44 | K>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs755394263 CA8591084 |
46 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399902354 rs1267725515 |
47 | V>L | No |
ClinGen gnomAD |
|
|
CA399902360 rs1290250512 |
48 | Q>* | No |
ClinGen TOPMed |
|
|
rs1293277255 CA399902363 |
48 | Q>R | No |
ClinGen Ensembl |
|
|
CA399902368 rs1451260759 |
49 | S>C | No |
ClinGen TOPMed |
|
| TCGA novel | 51 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs753200298 CA8591086 |
61 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8591085 rs779429695 |
61 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA399902458 rs1435830796 |
62 | V>E | No |
ClinGen TOPMed |
|
|
CA399902455 rs1173769761 |
62 | V>M | No |
ClinGen TOPMed |
|
|
CA8591087 rs758999530 |
63 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1188476998 CA399902510 |
71 | T>P | No |
ClinGen TOPMed |
|
|
rs1188476998 CA399902511 |
71 | T>S | No |
ClinGen TOPMed |
|
|
CA399902523 rs1371237650 |
73 | Q>E | No |
ClinGen gnomAD |
|
|
CA8591091 rs781253273 |
74 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA399902538 rs1598080208 |
75 | W>G | No |
ClinGen Ensembl |
|
|
CA8591093 rs769510993 |
77 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs769510993 CA399902553 |
77 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1469486227 CA399902562 |
78 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1469486227 CA399902563 |
78 | G>V | No |
ClinGen TOPMed gnomAD |
|
| rs758198899 | 80 | Q>A | Variant assessed as Somatic; 4.63e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768448857 CA8591097 |
81 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 84 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 86 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs924018125 CA290575640 |
88 | R>H | No |
ClinGen TOPMed |
|
|
rs11545626 CA290575643 |
89 | S>P | No |
ClinGen Ensembl |
|
|
CA290575645 rs1059968 VAR_028205 |
91 | T>N | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs1598080232 CA399902641 |
91 | T>P | No |
ClinGen Ensembl |
|
|
rs935334904 CA290575647 |
93 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1275817450 CA399902701 |
101 | V>M | No |
ClinGen gnomAD |
|
|
rs1323271014 CA399902713 |
102 | D>E | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 102 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399902724 rs955391996 |
104 | A>G | No |
ClinGen gnomAD |
|
|
CA8591099 rs761216623 |
104 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA290575652 rs955391996 |
104 | A>V | No |
ClinGen gnomAD |
|
|
CA399902737 rs1259002039 |
106 | A>G | No |
ClinGen gnomAD |
|
|
CA8591101 rs750033537 |
107 | E>* | No |
ClinGen ExAC gnomAD |
|
|
COSM3772929 rs765651568 CA8591103 |
108 | R>Q | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1267994847 CA399902747 |
108 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs753140896 CA8591104 |
113 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs758871820 CA8591105 |
113 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA399902788 rs1598080292 |
114 | V>G | No |
ClinGen Ensembl |
|
|
CA399902793 rs1363914035 |
115 | E>G | No |
ClinGen gnomAD |
|
|
rs372258038 CA8591107 |
116 | L>M | No |
ClinGen ESP ExAC gnomAD |
|
|
rs781336467 CA8591109 |
118 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA8591110 rs746029987 |
119 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399902829 rs770023977 |
121 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399902831 rs1290557660 |
121 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA8591111 rs770023977 |
121 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399902833 rs1350755959 |
122 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs868719891 CA290575668 |
123 | S>L | No |
ClinGen gnomAD |
|
|
rs1262743994 CA399902850 |
124 | D>E | No |
ClinGen TOPMed |
|
|
rs1188671534 CA399902871 |
127 | G>D | No |
ClinGen TOPMed |
|
|
CA399902880 rs1255574446 |
129 | P>T | No |
ClinGen TOPMed |
|
|
rs1050650948 CA290575680 |
136 | K>* | No |
ClinGen TOPMed gnomAD |
|
|
rs774152596 CA8591115 |
137 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA8591117 rs771429264 |
139 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA8591119 rs147354814 |
139 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399902956 CA8591121 rs776470066 |
141 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1423240165 CA399902980 |
145 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 149 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1252801023 CA399903023 |
151 | R>M | No |
ClinGen TOPMed |
|
|
rs763210222 CA8591122 |
151 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs764583641 CA8591123 |
153 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399903042 rs1598080375 |
154 | V>G | No |
ClinGen Ensembl |
|
|
CA8591124 rs751943750 |
155 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs757796175 CA8591125 |
155 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399903045 rs757796175 |
155 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399903056 rs1290641845 |
157 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1207138704 CA399903068 |
159 | A>T | No |
ClinGen gnomAD |
|
|
CA8591127 rs200026555 |
159 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs866011666 CA399903071 |
160 | A>P | No |
ClinGen TOPMed |
|
|
CA290575701 rs866011666 |
160 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA399903077 rs1300198511 |
161 | T>A | No |
ClinGen TOPMed |
|
|
rs377180469 CA8591128 |
162 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA290575704 rs1002576957 |
162 | L>P | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 162 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399903087 rs1184318605 |
163 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1384628575 CA399903091 |
163 | T>I | No |
ClinGen gnomAD |
|
|
CA399903088 rs1184318605 |
163 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
CA8591130 rs780243844 |
164 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA399903117 rs1415739288 |
167 | G>D | No |
ClinGen TOPMed |
|
| TCGA novel | 168 | C>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399903136 rs1461775427 |
170 | A>T | No |
ClinGen gnomAD |
|
|
rs896130940 CA290575714 |
173 | G>A | No |
ClinGen gnomAD |
|
|
CA8591132 rs754663381 |
173 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1335511948 CA399903170 |
175 | G>D | No |
ClinGen gnomAD |
|
|
rs1379853915 CA399903181 |
177 | Q>L | No |
ClinGen gnomAD |
|
|
rs1438284005 CA399903194 |
179 | G>S | No |
ClinGen TOPMed |
|
|
rs1298215920 CA399903214 |
182 | R>C | No |
ClinGen gnomAD |
|
|
rs1298215920 CA399903213 |
182 | R>G | No |
ClinGen gnomAD |
|
|
rs958350187 CA290575717 |
182 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA399903219 rs1225008880 |
183 | L>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1257494914 CA399903226 |
184 | Y>C | No |
ClinGen gnomAD |
|
| TCGA novel | 186 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399903275 rs61749921 |
190 | R>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA399903288 rs1204477694 |
192 | K>R | No |
ClinGen gnomAD |
|
|
COSM1290697 rs1044804021 CA290575722 |
195 | R>L | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA399903305 rs1044804021 |
195 | R>Q | No |
ClinGen TOPMed |
|
|
rs747991125 CA8591134 |
195 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1460025258 CA399903310 |
196 | G>D | No |
ClinGen gnomAD |
|
|
CA399903313 rs1191103589 |
197 | G>S | No |
ClinGen gnomAD |
|
| TCGA novel | 200 | R>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399903339 rs1168802757 |
200 | R>T | No |
ClinGen gnomAD |
No associated diseases with P49703
9 regional properties for P49703
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ribonuclease III domain | 1381 - 1610 | IPR000999-1 |
| domain | Ribonuclease III domain | 1643 - 1828 | IPR000999-2 |
| domain | Helicase, C-terminal | 371 - 542 | IPR001650 |
| domain | PAZ domain | 847 - 1027 | IPR003100 |
| domain | Dicer dimerisation domain | 571 - 667 | IPR005034 |
| domain | Helicase/UvrB, N-terminal | 12 - 182 | IPR006935 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 9 - 218 | IPR014001 |
| domain | Double-stranded RNA-binding domain | 1833 - 1896 | IPR014720 |
| domain | Dicer, double-stranded RNA-binding domain | 1832 - 1895 | IPR044441 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| protein secretion | The controlled release of proteins from a cell. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
48 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGNHLTEMAP | TASSFLPHFQ | ALHVVVIGLD | SAGKTSLLYR | LKFKEFVQSV | PTKGFNTEKI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RVPLGGSRGI | TFQVWDVGGQ | EKLRPLWRSY | TRRTDGLVFV | VDAAEAERLE | EAKVELHRIS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RASDNQGVPV | LVLANKQDQP | GALSAAEVEK | RLAVRELAAA | TLTHVQGCSA | VDGLGLQQGL |
| 190 | 200 | ||||
| ERLYEMILKR | KKAARGGKKR | R |