P49698
Gene name |
Hnf4a (Hnf-4, Hnf4, Nr2a1, Tcf14) |
Protein name |
Hepatocyte nuclear factor 4-alpha |
Names |
HNF-4-alpha, Nuclear receptor subfamily 2 group A member 1, Transcription factor 14, TCF-14, Transcription factor HNF-4 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:15378 |
EC number |
|
Protein Class |
|
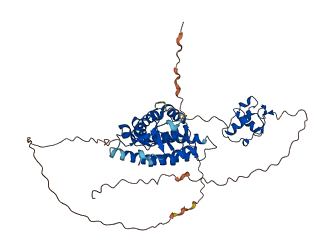
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P49698
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P49698-F1 | Predicted | AlphaFoldDB |
25 variants for P49698
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388595275 | 19 | L>M | No | EVA | |
| rs3388611522 | 30 | N>I | No | EVA | |
| rs3388595250 | 43 | S>C | No | EVA | |
| rs3388612965 | 63 | C>S | No | EVA | |
| rs3388613009 | 63 | C>Y | No | EVA | |
| rs3388612433 | 85 | R>K | No | EVA | |
| rs3392337486 | 132 | N>H | No | EVA | |
| rs3392501528 | 132 | N>I | No | EVA | |
| rs3388611531 | 144 | Y>* | No | EVA | |
| rs3388611028 | 160 | V>I | No | EVA | |
| rs3392535373 | 201 | W>C | No | EVA | |
| rs3392402073 | 203 | K>R | No | EVA | |
| rs3392435913 | 203 | K>T* | No | EVA | |
| rs3388602042 | 213 | L>P | No | EVA | |
| rs3388607997 | 239 | F>Y | No | EVA | |
| rs3388595233 | 266 | I>V | No | EVA | |
| rs3388612396 | 272 | L>P | No | EVA | |
| rs3388601365 | 274 | L>R | No | EVA | |
| rs3392402159 | 358 | I>V | No | EVA | |
| rs3392535312 | 362 | G>S | No | EVA | |
| rs3388607959 | 401 | N>K | No | EVA | |
| rs3388612356 | 405 | A>V | No | EVA | |
| rs3388612401 | 424 | R>K | No | EVA | |
| rs3388612407 | 425 | G>W | No | EVA | |
| rs3388611058 | 459 | P>T | No | EVA |
No associated diseases with P49698
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
23 GO annotations of molecular function
| Name | Definition |
|---|---|
| arachidonic acid binding | Binding to arachidonic acid, a straight chain fatty acid with 20 carbon atoms and four double bonds per molecule. Arachidonic acid is the all-Z-(5,8,11,14)-isomer. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by some RNA polymerase. The proximal promoter is in cis with and relatively close to the core promoter. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| fatty acid binding | Binding to a fatty acid, an aliphatic monocarboxylic acids liberated from naturally occurring fats and oils by hydrolysis. |
| fatty-acyl-CoA binding | Binding to a fatty-acyl-CoA, any derivative of coenzyme A in which the sulfhydryl group is in thiolester linkage with a fatty acyl group. |
| long-chain fatty acyl-CoA binding | Binding to a long-chain fatty acyl-CoA, any derivative of coenzyme A in which the sulfhydryl group is in a thioester linkage with a long-chain fatty-acyl group. Long-chain fatty-acyl-CoAs have chain lengths of C13 or more. |
| nuclear receptor activity | A DNA-binding transcription factor activity regulated by binding to a ligand that modulates the transcription of specific gene sets transcribed by RNA polymerase II. Nuclear receptor ligands are usually lipid-based (such as a steroid hormone) and the binding of the ligand to its receptor often occurs in the cytoplasm, which leads to its tranlocation to the nucleus. |
| palmitoyl-CoA hydrolase activity | Catalysis of the reaction: palmitoyl-CoA + H2O = CoA + palmitate. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II-specific DNA-binding transcription factor binding | Binding to a sequence-specific DNA binding RNA polymerase II transcription factor, any of the factors that interact selectively and non-covalently with a specific DNA sequence in order to modulate transcription. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| stearic acid binding | Binding to stearic acid, the 18-carbon saturated fatty acid octadecanoic acid. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
| transcription coregulator binding | Binding to a transcription coregulator, a protein involved in regulation of transcription via protein-protein interactions with transcription factors and other transcription regulatory proteins. Cofactors do not bind DNA directly, but rather mediate protein-protein interactions between regulatory transcription factors and the basal transcription machinery. |
| zinc ion binding | Binding to a zinc ion (Zn). |
42 GO annotations of biological process
| Name | Definition |
|---|---|
| acyl-CoA metabolic process | The chemical reactions and pathways involving acyl-CoA, any derivative of coenzyme A in which the sulfhydryl group is in thiolester linkage with an acyl group. |
| anatomical structure development | The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome. |
| blood coagulation | The sequential process in which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell-cell junction organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cell-cell junction. A cell-cell junction is a specialized region of connection between two cells. |
| cholesterol homeostasis | Any process involved in the maintenance of an internal steady state of cholesterol within an organism or cell. |
| establishment of tissue polarity | Coordinated organization of groups of cells in a tissue, such that they all orient to similar coordinates. |
| glucose homeostasis | Any process involved in the maintenance of an internal steady state of glucose within an organism or cell. |
| hepatocyte differentiation | The process in which a relatively unspecialized cell acquires the specialized features of a hepatocyte. A hepatocyte is specialized epithelial cell that is organized into interconnected plates called lobules, and is the main structural component of the liver. |
| lipid homeostasis | Any process involved in the maintenance of an internal steady state of lipid within an organism or cell. |
| lipid metabolic process | The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. |
| negative regulation of activation of Janus kinase activity | Any process that stops, prevents or reduces the frequency, rate or extent of activation of JAK (Janus Activated Kinase) kinase activity. |
| negative regulation of cell growth | Any process that stops, prevents, or reduces the frequency, rate, extent or direction of cell growth. |
| negative regulation of cell migration | Any process that stops, prevents, or reduces the frequency, rate or extent of cell migration. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of DNA-binding transcription factor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| negative regulation of mitotic cell cycle | Any process that stops, prevents or reduces the rate or extent of progression through the mitotic cell cycle. |
| negative regulation of protein import into nucleus | Any process that stops, prevents, or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus. |
| negative regulation of tyrosine phosphorylation of STAT protein | Any process that stops, prevents, or reduces the frequency, rate or extent of the introduction of a phosphate group to a tyrosine residue of a STAT (Signal Transducer and Activator of Transcription) protein. |
| ornithine metabolic process | The chemical reactions and pathways involving ornithine, an amino acid only rarely found in proteins, but which is important in living organisms as an intermediate in the reactions of the urea cycle and in arginine biosynthesis. |
| phospholipid homeostasis | Any process involved in the maintenance of an internal steady state of phospholipid within an organism or cell. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of fatty acid biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of fatty acids. |
| positive regulation of gluconeogenesis | Any process that activates or increases the frequency, rate or extent of gluconeogenesis. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of circadian rhythm | Any process that modulates the frequency, rate or extent of a circadian rhythm. A circadian rhythm is a biological process in an organism that recurs with a regularity of approximately 24 hours. |
| regulation of DNA-templated transcription | Any process that modulates the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of gastrulation | Any process that modulates the rate or extent of gastrulation. Gastrulation is the complex and coordinated series of cellular movements that occurs at the end of cleavage during embryonic development of most animals. |
| regulation of insulin secretion | Any process that modulates the frequency, rate or extent of the regulated release of insulin. |
| regulation of lipid metabolic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving lipids. |
| regulation of microvillus assembly | A process that modulates the formation of a microvillus. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| response to glucose | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| rhythmic process | Any process pertinent to the generation and maintenance of rhythms in the physiology of an organism. |
| sex differentiation | The establishment of the sex of an organism by physical differentiation. |
| signal transduction involved in regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression as a consequence of a process in which a signal is released and/or conveyed from one location to another. |
| SMAD protein signal transduction | The cascade of processes by which a signal interacts with a receptor, causing a change in the activity of a SMAD protein, and ultimately effecting a change in the functioning of the cell. |
| transcription by RNA polymerase II | The synthesis of RNA from a DNA template by RNA polymerase II (RNAP II), originating at an RNA polymerase II promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs). |
| triglyceride homeostasis | Any process involved in the maintenance of an internal steady state of triglyceride within an organism or cell. |
| type B pancreatic cell development | The process whose specific outcome is the progression of a type B pancreatic cell over time, from its formation to the mature structure. A type B pancreatic cell is a cell located towards center of the islets of Langerhans that secretes insulin. |
| xenobiotic metabolic process | The chemical reactions and pathways involving a xenobiotic compound, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P41235 | HNF4A | Hepatocyte nuclear factor 4-alpha | Homo sapiens (Human) | PR |
| P43135 | Nr2f2 | COUP transcription factor 2 | Mus musculus (Mouse) | PR |
| P43136 | Nr2f6 | Nuclear receptor subfamily 2 group F member 6 | Mus musculus (Mouse) | PR |
| P22449 | Hnf4a | Hepatocyte nuclear factor 4-alpha | Rattus norvegicus (Rat) | PR |
| Q21006 | nhr-34 | Nuclear hormone receptor family member nhr-34 | Caenorhabditis elegans | PR |
| Q21878 | nhr-1 | Nuclear hormone receptor family member nhr-1 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRLSKTLAGM | DMADYSAALD | PAYTTLEFEN | VQVLTMGNDT | SPSEGANLNS | SNSLGVSALC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AICGDRATGK | HYGASSCDGC | KGFFRRSVRK | NHMYSCRFSR | QCVVDKDKRN | QCRYCRLKKC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FRAGMKKEAV | QNERDRISTR | RSSYEDSSLP | SINALLQAEV | LSQQITSPIS | GINGDIRAKK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IANITDVCES | MKEQLLVLVE | WAKYIPAFCE | LLLDDQVALL | RAHAGEHLLL | GATKRSMVFK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DVLLLGNDYI | VPRHCPELAE | MSRVSIRILD | ELVLPFQELQ | IDDNEYACLK | AIIFFDPDAK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GLSDPGKIKR | LRSQVQVSLE | DYINDRQYDS | RGRFGELLLL | LPTLQSITWQ | MIEQIQFIKL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FGMAKIDNLL | QEMLLGGSAS | DAPHTHHPLH | PHLMQEHMGT | NVIVANTMPS | HLSNGQMCEW |
| 430 | 440 | 450 | 460 | 470 | |
| PRPRGQAATP | ETPQPSPPSG | SGSESYKLLP | GAITTIVKPP | SAIPQPTITK | QEAI |