P49137
Gene name |
MAPKAPK2 |
Protein name |
MAP kinase-activated protein kinase 2 |
Names |
MAPK-activated protein kinase 2 , MAPKAP kinase 2 , MAPKAP-K2 , MAPKAPK-2 , MK-2 , MK2 , EC 2.7.11.1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9261 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
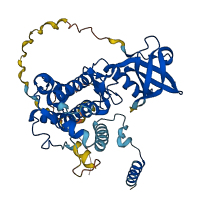
MAP KAP kinase 2 (MK2), a Ser/Thr kinase, plays a crucial role in the inflammatory process. MK2 contains an autoinhibitory domain within its C-terminal region. This domain regulates the enzyme by acting as a pseudosubstrate, blocking substrate binding and thus inhibiting activity. The autoinhibitory domain affects the catalytic kinase domain of MK2, specifically influencing the binding of peptide substrates and ATP, which are crucial for the enzyme’s activity. Phosphorylation by p38 MAP kinase alleviates autoinhibition. It induces conformational changes in MK2, particularly in the autoinhibitory domain and the activation segment, allowing efficient substrate binding and restoring enzymatic activity.
Autoinhibitory domains (AIDs)
Target domain |
5-325 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis, Deletion assay |
Accessory elements
206-228 (Activation loop from InterPro)
Target domain |
64-325 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Seternes OM et al. (2002) "Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress", Molecular and cellular biology, 22, 6931-45
- Engel K et al. (1998) "Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation", The EMBO journal, 17, 3363-71
Autoinhibited structure
Activated structure

30 structures for P49137
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1KWP | X-ray | 280 A | A/B | 1-400 | PDB |
| 1NXK | X-ray | 270 A | A/B/C/D | 1-400 | PDB |
| 1NY3 | X-ray | 300 A | A | 1-400 | PDB |
| 2JBO | X-ray | 310 A | A | 41-364 | PDB |
| 2JBP | X-ray | 331 A | A/B/C/D/E/F/G/H/I/J/K/L | 41-364 | PDB |
| 2OKR | X-ray | 200 A | C/F | 370-393 | PDB |
| 2ONL | X-ray | 400 A | C/D | 1-400 | PDB |
| 2OZA | X-ray | 270 A | A | 47-400 | PDB |
| 2P3G | X-ray | 380 A | X | 45-371 | PDB |
| 2PZY | X-ray | 290 A | A/B/C/D | 41-364 | PDB |
| 3A2C | X-ray | 290 A | A/B/C/D/E/F/G/H/I/J/K/L | 41-364 | PDB |
| 3FPM | X-ray | 330 A | A | 41-364 | PDB |
| 3FYJ | X-ray | 380 A | X | 45-371 | PDB |
| 3FYK | X-ray | 350 A | X | 45-371 | PDB |
| 3GOK | X-ray | 320 A | A/B/C/D/E/F/G/H/I/J/K/L | 41-364 | PDB |
| 3KA0 | X-ray | 290 A | A | 47-366 | PDB |
| 3KC3 | X-ray | 290 A | A/B/C/D/E/F/G/H/I/J/K/L | 41-364 | PDB |
| 3KGA | X-ray | 255 A | A | 47-364 | PDB |
| 3M2W | X-ray | 241 A | A | 47-364 | PDB |
| 3M42 | X-ray | 268 A | A | 47-364 | PDB |
| 3R2B | X-ray | 290 A | A/B/C/D/E/F/G/H/I/J/K/L | 47-364 | PDB |
| 3R2Y | X-ray | 300 A | A | 46-364 | PDB |
| 3R30 | X-ray | 320 A | A | 46-364 | PDB |
| 3WI6 | X-ray | 299 A | A/B/C/D/E/F | 41-364 | PDB |
| 4TYH | X-ray | 300 A | A | 51-400 | PDB |
| 6T8X | X-ray | 281 A | A/B/C/D/E/F | 41-338 | PDB |
| 6TCA | X-ray | 370 A | A/C/E/G | 41-400 | PDB |
| 7NRY | X-ray | 380 A | X | 45-371 | PDB |
| 8XU4 | X-ray | 340 A | A/B/C/D/E/F/G/H/I/J/K/L | 47-364 | PDB |
| AF-P49137-F1 | Predicted | AlphaFoldDB |
225 variants for P49137
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001262851 rs1673811124 |
149 | R>* | Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar dbSNP |
|
CA344484263 rs1553425290 |
3 | S>P | No |
ClinGen Ensembl |
|
|
rs1245550536 CA344484308 |
5 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1245550536 CA344484302 |
5 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1284284124 CA344484353 |
7 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1553425301 CA344484368 |
8 | Q>R | No |
ClinGen Ensembl |
|
|
CA344484410 rs1206536591 |
10 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA344484407 rs1206536591 |
10 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA344484435 rs1308290172 |
12 | V>M | No |
ClinGen TOPMed |
|
|
rs1246509011 CA344484460 |
13 | P>L | No |
ClinGen TOPMed |
|
|
rs1246509011 CA344484457 |
13 | P>Q | No |
ClinGen TOPMed |
|
|
CA344484458 rs1246509011 |
13 | P>R | No |
ClinGen TOPMed |
|
| TCGA novel | 14 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA344484483 rs1319019572 |
15 | P>R | No |
ClinGen TOPMed |
|
|
CA344484479 rs1553425319 |
15 | P>S | No |
ClinGen gnomAD |
|
|
CA344484489 rs1553425321 |
16 | A>P | No |
ClinGen Ensembl |
|
|
CA344484494 rs1553425322 |
16 | A>V | No |
ClinGen gnomAD |
|
|
CA344484521 rs1553425325 |
18 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 19 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1553425339 CA344484547 |
21 | P>S | No |
ClinGen gnomAD |
|
|
CA344484560 rs1425759842 |
22 | Q>L | No |
ClinGen TOPMed gnomAD |
|
|
CA344484562 rs1425759842 |
22 | Q>P | No |
ClinGen TOPMed gnomAD |
|
|
rs782036969 CA1363301 |
23 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363302 rs782186251 |
24 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363305 rs181345810 |
25 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 25 | T>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs181345810 CA1363306 |
25 | T>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA344484606 rs1558569683 |
26 | P>H | No |
ClinGen Ensembl |
|
|
rs1553425349 CA344484613 |
27 | A>P | No |
ClinGen gnomAD |
|
|
CA344484628 rs1553425354 |
28 | L>P | No |
ClinGen gnomAD |
|
|
rs782139014 CA1363309 |
30 | H>P | No |
ClinGen ExAC |
|
|
CA344484653 rs782797008 CA1363310 |
30 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363312 rs782064315 |
32 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1338511158 CA344484664 |
32 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA344484662 rs1338511158 |
32 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA344484668 rs573218643 |
33 | A>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1363313 rs573218643 |
33 | A>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA344484666 rs1553425370 |
33 | A>P | No |
ClinGen gnomAD |
|
|
CA344484672 rs1553425374 |
34 | Q>E | No |
ClinGen gnomAD |
|
|
CA344484680 rs1572470610 |
35 | P>S | No |
ClinGen Ensembl |
|
|
CA1363319 rs782504552 |
37 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1553425392 CA344484699 |
38 | P>L | No |
ClinGen gnomAD |
|
|
CA1363320 rs782674769 |
38 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1553425396 CA344484700 |
39 | P>S | No |
ClinGen gnomAD |
|
|
CA344484707 rs1572470680 |
40 | P>A | No |
ClinGen Ensembl |
|
|
CA1363322 rs782451973 |
40 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782451973 CA344484710 |
40 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA344484714 rs1553425404 |
41 | Q>L | No |
ClinGen gnomAD |
|
|
rs782578871 CA1363323 |
43 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1244786492 CA344484757 |
44 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA344484753 rs1244786492 |
44 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA344484755 rs1244786492 |
44 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA344484801 rs782364714 |
47 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1275111539 CA344484847 |
51 | G>C | No |
ClinGen TOPMed |
|
|
rs146073998 COSM4143283 COSM4143282 CA1363327 |
51 | G>V | thyroid [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs370656530 CA1363328 |
53 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1553425419 CA344484879 |
54 | I>V | No |
ClinGen gnomAD |
|
|
rs1277577091 CA344484889 |
55 | K>Q | No |
ClinGen TOPMed |
|
|
CA344484931 rs1553425427 |
58 | A>T | No |
ClinGen gnomAD |
|
|
rs1553425429 CA344484941 |
58 | A>V | No |
ClinGen gnomAD |
|
|
rs782041388 CA1363330 |
60 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA1363335 rs781888295 |
68 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA344485199 rs1436257745 |
72 | L>P | No |
ClinGen TOPMed |
|
|
rs1331532151 CA344485279 |
75 | N>S | No |
ClinGen TOPMed |
|
|
rs1553425445 CA344485435 |
83 | N>S | No |
ClinGen gnomAD |
|
|
rs1572470877 CA344485454 |
84 | K>R | No |
ClinGen Ensembl |
|
| TCGA novel | 84 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 85 | R>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1422330364 CA344485474 |
85 | R>S | No |
ClinGen TOPMed |
|
| TCGA novel | 87 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1553425447 CA344485497 |
87 | Q>P | No |
ClinGen gnomAD |
|
|
rs781807175 CA1363338 |
91 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA344485581 rs782468736 |
92 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs782468736 CA1363339 |
92 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1553432180 CA344476243 |
94 | M>I | No |
ClinGen gnomAD |
|
|
CA344476348 rs1553432187 |
101 | A>S | No |
ClinGen gnomAD |
|
| TCGA novel | 102 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM2156166 rs151079567 COSM2156165 CA1363361 |
102 | R>H | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs151079567 CA1363362 |
102 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA344476452 rs1553432196 |
108 | H>Q | No |
ClinGen gnomAD |
|
|
CA344476463 rs1166739803 |
109 | W>L | No |
ClinGen TOPMed |
|
|
rs782527754 CA1363366 |
110 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363365 rs375800332 |
110 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782185154 CA1363368 |
111 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA1363370 rs374754928 |
113 | Q>H | No |
ClinGen ESP ExAC gnomAD |
|
|
CA344476516 rs868968219 |
113 | Q>P | No |
ClinGen gnomAD |
|
|
CA1363371 rs782231165 |
115 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1553432206 CA344476558 |
116 | H>R | No |
ClinGen gnomAD |
|
|
rs782152248 CA1363375 |
118 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs141946496 CA1363376 |
119 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA344476599 rs1553432209 |
119 | R>W | No |
ClinGen gnomAD |
|
|
CA344476610 rs1553432211 |
120 | I>V | No |
ClinGen gnomAD |
|
|
rs781915750 CA1363377 |
121 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA344476645 rs1572515040 |
122 | D>G | No |
ClinGen Ensembl |
|
|
rs1553432214 CA344476652 |
123 | V>M | No |
ClinGen gnomAD |
|
|
rs534203474 CA36555664 |
127 | L>V | No |
ClinGen Ensembl |
|
|
COSM902999 COSM1583988 CA344476742 rs1363430228 |
129 | A>T | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs781887326 CA1363383 |
130 | G>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 131 | R>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782432987 CA1363384 |
131 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs886806296 CA36555740 |
133 | C>Y | No |
ClinGen Ensembl |
|
|
rs1553432226 CA344476887 |
140 | C>S | No |
ClinGen gnomAD |
|
|
rs1347942859 CA344476953 |
141 | L>M | No |
ClinGen TOPMed |
|
|
rs782082445 CA1363404 |
141 | L>S | No |
ClinGen ExAC gnomAD |
|
|
rs1311181554 CA344476992 |
143 | G>D | No |
ClinGen TOPMed |
|
|
CA344476982 rs1230603507 |
143 | G>S | No |
ClinGen TOPMed |
|
| TCGA novel | 145 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1363406 rs781851372 |
148 | S>G | No |
ClinGen ExAC TOPMed |
|
|
CA344477051 rs1348592671 |
148 | S>N | No |
ClinGen TOPMed |
|
|
rs782516721 CA1363407 |
149 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 151 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA344477101 rs1376132455 |
152 | D>A | No |
ClinGen TOPMed |
|
|
CA36555958 rs938576272 |
153 | R>G | No |
ClinGen Ensembl |
|
|
rs1175840148 CA344477138 |
155 | D>E | No |
ClinGen TOPMed |
|
|
CA1363409 rs527268461 |
156 | Q>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1927707 CA36556348 COSM275991 rs772956044 |
165 | E>K | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA344477842 rs1405298202 |
167 | M>V | No |
ClinGen TOPMed |
|
|
CA344477886 rs1397768555 |
170 | I>V | No |
ClinGen TOPMed |
|
|
CA344477896 rs1553432317 |
171 | G>S | No |
ClinGen gnomAD |
|
|
VAR_040753 rs35671930 CA1363436 |
173 | A>G | No |
ClinGen UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1173858137 CA344477950 |
175 | Q>* | No |
ClinGen TOPMed |
|
|
rs1553432321 CA344477968 |
176 | Y>C | No |
ClinGen gnomAD |
|
| TCGA novel | 180 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA344478008 rs1478924734 |
180 | I>V | No |
ClinGen TOPMed |
|
|
CA1363440 rs149638565 |
182 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1043183855 CA36556389 |
185 | R>Q | No |
ClinGen gnomAD |
|
|
CA36556737 rs561595271 |
194 | Y>C | No |
ClinGen Ensembl |
|
|
CA1363462 rs531894220 |
195 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA344478256 rs531894220 |
195 | T>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1363464 rs782384462 |
197 | K>R | No |
ClinGen ExAC |
|
|
CA344478328 rs375671223 |
200 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA344478322 rs1553432393 |
200 | N>S | No |
ClinGen gnomAD |
|
|
CA1363466 rs782149296 |
201 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs140901506 CA1363467 |
202 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA344478344 rs1484281453 |
202 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs368050217 CA1363469 |
206 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782803587 CA1363475 |
212 | K>Q | No |
ClinGen ExAC |
|
|
CA1363476 rs74538056 |
213 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs782769109 CA36556910 |
214 | T>I | No |
ClinGen gnomAD |
|
|
CA1363477 rs571837891 |
214 | T>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1363478 rs200028198 |
215 | T>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1257277521 CA344478523 |
216 | S>T | No |
ClinGen TOPMed |
|
|
CA344478535 rs1230915063 |
217 | H>R | No |
ClinGen TOPMed |
|
|
CA1363479 rs781830804 |
218 | N>D | No |
ClinGen ExAC |
|
| TCGA novel | 229 | Y>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA344478938 rs1261797849 |
243 | S>C | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 244 | C>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782414991 CA1363508 |
246 | M>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 249 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA344479040 rs1203635358 |
251 | V>I | No |
ClinGen TOPMed |
|
|
rs375137265 CA1363510 |
252 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1572517856 CA344479205 |
260 | Y>D | No |
ClinGen Ensembl |
|
| TCGA novel | 263 | F>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1553432591 | 263 | F>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1363529 rs782375243 |
266 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363530 rs782008886 |
266 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA344479287 rs782008886 |
266 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781923393 CA1363533 |
271 | I>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 272 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1553432605 CA344479434 |
278 | R>G | No |
ClinGen gnomAD |
|
|
COSM1286098 COSM1286099 rs1553432606 CA344479439 |
278 | R>H | Variant assessed as Somatic; 0.0 impact. autonomic_ganglia [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1247727262 CA344479456 |
280 | R>G | No |
ClinGen TOPMed |
|
|
rs782072138 CA1363534 COSM3771605 COSM3771606 |
280 | R>Q | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA344479474 rs1483324985 |
281 | M>K | No |
ClinGen TOPMed |
|
|
CA1363536 rs781968571 |
283 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs782729938 CA1363535 |
283 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA1363538 rs142063401 |
288 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1553432613 CA344479613 |
291 | W>C | No |
ClinGen gnomAD |
|
| TCGA novel | 292 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1261683933 CA344480344 |
300 | M>L | No |
ClinGen TOPMed |
|
|
CA344480343 rs1261683933 |
300 | M>V | No |
ClinGen TOPMed |
|
|
CA1363572 rs545600073 |
303 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1280991127 CA344480384 |
303 | R>W | No |
ClinGen TOPMed |
|
|
CA344480399 rs1334705583 |
304 | N>S | No |
ClinGen TOPMed |
|
|
rs1558589974 CA344480474 |
311 | T>A | No |
ClinGen Ensembl |
|
|
CA1363574 rs782597489 |
311 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1363573 rs782597489 |
311 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA344480537 rs1454535420 |
316 | I>V | No |
ClinGen TOPMed |
|
|
rs868917636 CA344480560 |
318 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs1553432730 CA344480567 |
318 | E>V | No |
ClinGen gnomAD |
|
|
rs782279440 CA1363577 |
320 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782279440 CA344480609 |
320 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 322 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 324 | W>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1553432736 CA344480695 |
326 | M>T | No |
ClinGen gnomAD |
|
|
rs782425128 CA1363601 |
327 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs1292139600 CA344480749 |
327 | Q>R | No |
ClinGen TOPMed |
|
|
rs782072519 CA1363603 |
328 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA1363604 rs782340351 |
329 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA36558283 rs970977724 |
330 | K>M | No |
ClinGen TOPMed |
|
|
rs868952591 CA344480807 |
332 | P>T | No |
ClinGen Ensembl |
|
|
rs1363748424 CA344480817 |
333 | Q>E | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 339 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs941786201 CA344480907 |
340 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs941786201 CA36558301 |
340 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs142025224 CA1363609 |
340 | R>W | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1553432760 CA344480951 |
344 | E>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA344481004 rs782749899 |
348 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA1363613 rs782749899 |
348 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1553432763 CA344481002 |
348 | R>W | No |
ClinGen gnomAD |
|
|
rs140988938 CA1363614 |
353 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1572519960 CA344481230 |
360 | L>S | No |
ClinGen Ensembl |
|
|
VAR_040754 CA1363667 rs55894011 |
361 | A>S | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs782118884 CA1363668 |
362 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA344481253 rs1553432908 |
364 | R>C | No |
ClinGen gnomAD |
|
|
rs782022011 COSM903001 CA1363670 |
365 | V>I | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs782704000 CA1363672 |
368 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA344481326 rs1553432914 |
374 | K>E | No |
ClinGen gnomAD |
|
|
CA36559009 rs892611562 |
374 | K>M | No |
ClinGen TOPMed |
|
| rs1553432912 | 374 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1337144318 CA344481336 |
375 | I>T | No |
ClinGen TOPMed |
|
|
rs781818804 CA1363673 |
377 | D>V | No |
ClinGen ExAC |
|
|
CA344481370 rs1553432918 |
380 | N>S | No |
ClinGen gnomAD |
|
| TCGA novel | 386 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1572520041 CA344481412 |
387 | R>Q | No |
ClinGen Ensembl |
|
|
CA1363678 rs782678368 |
390 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs191175287 CA1363679 |
391 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA344481437 rs1553432921 |
391 | R>W | No |
ClinGen gnomAD |
|
|
rs1558590699 CA344481443 COSM4143286 |
392 | A>D | thyroid [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA1363681 rs782600274 |
394 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA1363682 rs782232686 |
396 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA1363684 rs782652551 |
397 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 397 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782282911 CA1363685 |
398 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781933066 CA1363687 |
400 | H>Q | No |
ClinGen ExAC gnomAD |
No associated diseases with P49137
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein serine/threonine kinase activity | Calcium-dependent catalysis of the reactions |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions |
| mitogen-activated protein kinase binding | Binding to a mitogen-activated protein kinase. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
23 GO annotations of biological process
| Name | Definition |
|---|---|
| 3'-UTR-mediated mRNA stabilization | An mRNA stabilization process in which one or more RNA-binding proteins associate with the 3'-untranslated region (UTR) of an mRNA. |
| cellular response to vascular endothelial growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vascular endothelial growth factor stimulus. |
| DNA damage response | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| inner ear development | The process whose specific outcome is the progression of the inner ear over time, from its formation to the mature structure. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| leukotriene metabolic process | The chemical reactions and pathways involving leukotriene, a pharmacologically active substance derived from a polyunsaturated fatty acid, such as arachidonic acid. |
| macropinocytosis | An endocytosis process that results in the uptake of liquid material by cells from their external environment by the 'ruffling' of the cell membrane to form heterogeneously sized intracellular vesicles called macropinosomes, which can be up to 5 micrometers in size. |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers |
| p38MAPK cascade | An intracellular protein kinase cascade containing at least a p38 MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tier |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of macrophage cytokine production | Any process that increases the rate, frequency or extent of macrophage cytokine production. Macrophage cytokine production is the appearance of a chemokine due to biosynthesis or secretion following a cellular stimulus, resulting in an increase in its intracellular or extracellular levels. |
| positive regulation of tumor necrosis factor production | Any process that activates or increases the frequency, rate or extent of tumor necrosis factor production. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cellular response to heat | Any process that modulates the frequency, rate or extent of cellular response to heat. |
| regulation of interleukin-6 production | Any process that modulates the frequency, rate, or extent of interleukin-6 production. |
| regulation of mRNA stability | Any process that modulates the propensity of mRNA molecules to degradation. Includes processes that both stabilize and destabilize mRNAs. |
| regulation of tumor necrosis factor production | Any process that modulates the frequency, rate or extent of tumor necrosis factor production. |
| regulation of tumor necrosis factor-mediated signaling pathway | Any process that modulates the rate or extent of the tumor necrosis factor-mediated signaling pathway. The tumor necrosis factor-mediated signaling pathway is the series of molecular signals generated as a consequence of tumor necrosis factor binding to a cell surface receptor. |
| response to cytokine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor of a target cell. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
| vascular endothelial growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a vascular endothelial growth factor receptor (VEGFR) on the surface of the target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3SYZ2 | MAPKAPK3 | MAP kinase-activated protein kinase 3 | Bos taurus (Bovine) | SS |
| P49071 | MAPk-Ak2 | MAP kinase-activated protein kinase 2 | Drosophila melanogaster (Fruit fly) | SS |
| Q8IW41 | MAPKAPK5 | MAP kinase-activated protein kinase 5 | Homo sapiens (Human) | SS |
| Q16644 | MAPKAPK3 | MAP kinase-activated protein kinase 3 | Homo sapiens (Human) | SS |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| O54992 | Mapkapk5 | MAP kinase-activated protein kinase 5 | Mus musculus (Mouse) | EV |
| Q3UMW7 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Mus musculus (Mouse) | SS |
| P49138 | Mapkapk2 | MAP kinase-activated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q66H84 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Rattus norvegicus (Rat) | SS |
| Q965G5 | mak-2 | MAP kinase-activated protein kinase mak-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLSNSQGQSP | PVPFPAPAPP | PQPPTPALPH | PPAQPPPPPP | QQFPQFHVKS | GLQIKKNAII |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DDYKVTSQVL | GLGINGKVLQ | IFNKRTQEKF | ALKMLQDCPK | ARREVELHWR | ASQCPHIVRI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VDVYENLYAG | RKCLLIVMEC | LDGGELFSRI | QDRGDQAFTE | REASEIMKSI | GEAIQYLHSI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NIAHRDVKPE | NLLYTSKRPN | AILKLTDFGF | AKETTSHNSL | TTPCYTPYYV | APEVLGPEKY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DKSCDMWSLG | VIMYILLCGY | PPFYSNHGLA | ISPGMKTRIR | MGQYEFPNPE | WSEVSEEVKM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LIRNLLKTEP | TQRMTITEFM | NHPWIMQSTK | VPQTPLHTSR | VLKEDKERWE | DVKEEMTSAL |
| 370 | 380 | 390 | |||
| ATMRVDYEQI | KIKKIEDASN | PLLLKRRKKA | RALEAAALAH |