P46109
Gene name |
CRKL |
Protein name |
Crk-like protein |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1399 |
EC number |
|
Protein Class |
|
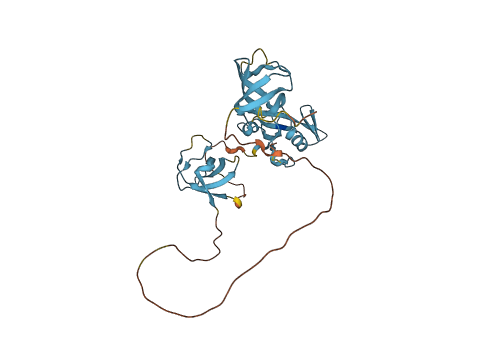
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
123-183 (N-terminal SH3 domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Kobashigawa Y et al. (2007) "Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK", Nature structural & molecular biology, 14, 503-10
- Ogawa S et al. (1994) "The C-terminal SH3 domain of the mouse c-Crk protein negatively regulates tyrosine-phosphorylation of Crk associated p130 in rat 3Y1 cells", Oncogene, 9, 1669-78
- Sriram G et al. (2012) "Commentary: The carboxyl-terminal Crk SH3 domain: Regulatory strategies and new perspectives", FEBS letters, 586, 2615-8
- Takeuchi K et al. (2010) "Autoinhibitory interaction in the multidomain adaptor protein Nck: possible roles in improving specificity and functional diversity", Biochemistry, 49, 5634-41
Autoinhibited structure

Activated structure

171 variants for P46109
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA10117262 rs745921293 |
2 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA410765205 rs1463139007 |
3 | S>F | No |
ClinGen gnomAD |
|
|
CA410765207 rs1378084235 |
4 | A>T | No |
ClinGen TOPMed |
|
|
rs1394048815 CA410765212 |
4 | A>V | No |
ClinGen gnomAD |
|
|
rs1467146233 CA410765213 |
5 | R>G | No |
ClinGen gnomAD |
|
|
CA410765218 rs778958922 |
5 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10117264 rs778958922 |
5 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1388023973 CA410765236 |
8 | S>P | No |
ClinGen TOPMed |
|
|
CA410765246 COSM1751748 rs1430434565 |
9 | S>L | urinary_tract [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs547899646 CA10117266 |
10 | D>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs567876179 CA10117267 |
11 | R>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs567876179 CA322296275 |
11 | R>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1458100931 CA410765259 |
12 | S>P | No |
ClinGen TOPMed |
|
|
rs770480792 CA10117269 |
13 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs776257196 CA10117270 |
13 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs865927265 CA322296294 |
16 | M>V | No |
ClinGen Ensembl |
|
|
CA10117271 rs758939601 |
17 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs138882903 CA322296307 |
18 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10117272 rs138882903 |
18 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1180503338 CA410765306 |
19 | V>G | No |
ClinGen TOPMed |
|
|
CA10117273 rs775929456 |
19 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA10117274 rs763322278 |
22 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA322296324 rs62238487 |
22 | Q>K | No |
ClinGen Ensembl |
|
|
CA410765330 rs1460095339 |
23 | E>G | No |
ClinGen gnomAD |
|
|
CA322296351 rs888463667 |
26 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA10117276 rs751864009 |
26 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA410765350 rs751864009 |
26 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1601669816 CA410765365 |
29 | Q>P | No |
ClinGen Ensembl |
|
|
rs756219565 CA10117280 |
30 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs199641129 CA322296404 |
32 | R>L | No |
ClinGen Ensembl |
|
|
rs770661007 CA10117286 |
35 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746762587 CA10117285 |
35 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs746762587 CA410765402 |
35 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs1601669837 CA410765419 |
37 | L>F | No |
ClinGen Ensembl |
|
|
CA410765431 rs1422177625 |
39 | R>G | No |
ClinGen gnomAD |
|
|
CA322296472 rs966859460 |
39 | R>H | No |
ClinGen Ensembl |
|
|
rs1364659560 CA410765435 |
40 | D>H | No |
ClinGen gnomAD |
|
|
rs1601669855 CA410765454 |
43 | T>S | No |
ClinGen Ensembl |
|
|
rs769136802 CA10117292 |
44 | C>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1157261023 CA410765463 |
44 | C>Y | No |
ClinGen gnomAD |
|
|
CA10117293 rs774827798 |
45 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1236385760 CA410765547 |
57 | R>Q | No |
ClinGen gnomAD |
|
|
rs1364651927 CA410765568 |
59 | S>C | No |
ClinGen gnomAD |
|
|
CA410765594 rs1278191120 |
61 | Y>C | No |
ClinGen gnomAD |
|
|
CA10117298 rs766496631 |
64 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1418708599 CA410765655 |
65 | S>L | No |
ClinGen TOPMed |
|
|
CA10117299 rs753956143 |
67 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA322296521 rs973151946 |
68 | N>H | No |
ClinGen Ensembl |
|
|
CA410765687 rs1397083945 |
68 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1463918076 CA410765838 |
76 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs745447266 CA10117305 |
77 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA10117307 rs779594981 |
80 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1327554664 CA410765964 |
82 | P>L | No |
ClinGen gnomAD |
|
|
CA322296590 rs1054393986 |
82 | P>S | No |
ClinGen Ensembl |
|
|
rs3180409 CA322296595 |
91 | H>Y | No |
ClinGen Ensembl |
|
|
CA410766131 rs1338868128 |
92 | Y>C | No |
ClinGen gnomAD |
|
|
rs773484875 CA10117313 |
97 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1257146277 CA410766256 |
99 | I>L | No |
ClinGen TOPMed |
|
|
CA410766262 rs1257146277 |
99 | I>V | No |
ClinGen TOPMed |
|
|
CA410766275 rs1569130085 |
100 | E>Q | No |
ClinGen Ensembl |
|
|
CA10117316 rs776895332 |
103 | P>H | No |
ClinGen ExAC gnomAD |
|
|
rs766688775 CA10117315 |
103 | P>S | No |
ClinGen ExAC |
|
|
rs1399022200 CA410766342 |
104 | R>G | No |
ClinGen TOPMed |
|
|
CA410761882 rs1213587964 |
106 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA410761884 rs775637595 |
106 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA10117337 rs775637595 |
106 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs761713297 CA10117338 |
107 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10117340 rs767466740 |
107 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs755863490 CA10117341 |
108 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA410761899 rs1188009275 |
109 | P>S | No |
ClinGen TOPMed |
|
|
CA10117342 rs200265850 |
110 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA10117343 rs753427288 COSM1751749 |
114 | S>L | urinary_tract [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA410761936 rs1378195752 |
115 | A>S | No |
ClinGen gnomAD |
|
|
rs925122172 CA322269072 |
115 | A>V | No |
ClinGen Ensembl |
|
|
rs376691633 CA10117344 |
116 | P>T | No |
ClinGen ESP ExAC gnomAD |
|
|
CA10117345 rs778350864 |
117 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs936540793 CA322269077 |
117 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA322269075 rs3180410 |
117 | N>Y | No |
ClinGen Ensembl |
|
|
rs1451933601 CA410761961 |
120 | T>A | No |
ClinGen Ensembl |
|
|
rs1447468704 CA410761965 |
120 | T>I | No |
ClinGen gnomAD |
|
|
CA410761968 rs1301110218 |
121 | A>T | No |
ClinGen gnomAD |
|
|
CA10117346 rs752232310 |
122 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs148491827 CA322269082 |
122 | E>K | No |
ClinGen Ensembl |
|
|
rs1276902404 CA410761986 |
124 | N>H | No |
ClinGen TOPMed |
|
|
CA410761990 rs1220289351 |
124 | N>S | No |
ClinGen TOPMed |
|
|
CA410761999 rs1344311195 |
126 | E>K | No |
ClinGen gnomAD |
|
|
rs758872714 CA10117347 |
127 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1309018634 CA410762030 |
130 | T>S | No |
ClinGen gnomAD |
|
|
rs1221209545 CA410762032 COSM579733 |
131 | L>V | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA10117348 rs778388431 |
137 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs747404847 CA10117349 |
139 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747404847 CA410762090 |
139 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781570771 CA10117351 |
143 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs770045446 CA10117353 |
145 | K>E | No |
ClinGen ExAC |
|
|
rs775479004 CA10117354 |
146 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA10117356 rs771933873 |
152 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1276223674 CA410762284 |
153 | I>T | No |
ClinGen gnomAD |
|
|
CA410762496 rs1235946709 |
162 | S>N | No |
ClinGen gnomAD |
|
|
CA410762531 rs760471921 |
164 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA10117359 rs200712124 |
164 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10117358 rs760471921 |
164 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs373785511 CA322269138 |
169 | R>Q | No |
ClinGen ESP TOPMed |
|
|
CA410762620 rs1356309955 |
169 | R>W | No |
ClinGen gnomAD |
|
|
rs759166693 CA10117361 |
175 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs535421669 CA10117362 |
178 | V>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1217926975 CA410762779 |
179 | E>G | No |
ClinGen gnomAD |
|
|
CA410762771 rs1260646700 |
179 | E>K | No |
ClinGen Ensembl |
|
|
rs1307610334 CA410762804 |
181 | L>F | No |
ClinGen gnomAD |
|
|
rs952900119 CA322269152 |
181 | L>P | No |
ClinGen Ensembl |
|
|
CA410762814 rs1253813166 |
182 | V>L | No |
ClinGen TOPMed |
|
|
rs1253813166 CA410762811 |
182 | V>M | No |
ClinGen TOPMed |
|
|
CA410762884 rs1451107236 |
185 | S>L | No |
ClinGen TOPMed |
|
|
rs147576993 CA10117366 |
186 | P>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA322269176 rs147576993 |
186 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs757836194 CA10117367 |
187 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA10117369 rs746219908 |
188 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA322269196 rs199907840 |
190 | H>R | No |
ClinGen 1000Genomes TOPMed |
|
|
rs1267925221 CA410762971 |
192 | N>S | No |
ClinGen TOPMed |
|
|
CA322269203 rs748247058 |
196 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA10117372 rs749349384 |
197 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA410763061 rs1569134891 |
202 | E>K | No |
ClinGen Ensembl |
|
|
CA10117375 rs200629140 |
203 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA10117376 rs770839945 |
204 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1601679635 CA410763076 |
204 | A>V | No |
ClinGen Ensembl |
|
|
CA322269260 COSM1415033 rs976691655 |
205 | H>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1363256176 CA410763133 |
213 | T>A | No |
ClinGen gnomAD |
|
|
rs1323862187 CA410763143 |
214 | T>I | No |
ClinGen TOPMed |
|
|
rs1336540725 CA410763149 |
215 | P>L | No |
ClinGen gnomAD |
|
|
rs764823179 CA10117380 |
217 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1479479199 CA410763162 |
218 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA10117382 rs762375550 |
218 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA410763169 rs1385927465 |
219 | V>A | No |
ClinGen TOPMed |
|
|
rs145813322 CA10117384 |
221 | G>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs757890975 CA10117385 |
221 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768104683 CA10117386 |
222 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1421511528 CA410763188 |
223 | P>S | No |
ClinGen gnomAD |
|
|
CA10117387 rs750783216 |
225 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1165973049 CA410763203 |
226 | A>T | No |
ClinGen gnomAD |
|
|
CA10117388 rs756451346 |
226 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA410763209 rs1404011600 |
227 | I>V | No |
ClinGen gnomAD |
|
|
CA410763216 rs1477769854 |
228 | T>A | No |
ClinGen TOPMed |
|
|
CA410763215 rs1477769854 |
228 | T>P | No |
ClinGen TOPMed |
|
|
rs1320625946 CA410763227 |
230 | L>M | No |
ClinGen gnomAD |
|
|
rs1601679701 CA410763244 |
232 | S>C | No |
ClinGen Ensembl |
|
|
CA10117389 rs780153751 |
233 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA410763262 rs1201043851 |
235 | N>T | No |
ClinGen TOPMed |
|
|
rs1469767752 CA410763286 |
239 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs749456958 CA10117390 |
240 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA10117392 rs557938965 |
243 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1233334173 CA410763322 |
244 | Q>R | No |
ClinGen gnomAD |
|
|
CA410763357 rs1283293456 |
248 | P>T | No |
ClinGen gnomAD |
|
|
CA410763475 rs1284642010 |
257 | A>P | No |
ClinGen gnomAD |
|
|
rs1184315163 CA410765848 |
263 | I>T | No |
ClinGen gnomAD |
|
|
CA322278252 rs564684352 |
266 | V>I | No |
ClinGen 1000Genomes gnomAD |
|
|
rs138367739 CA322278271 |
268 | R>K | No |
ClinGen ESP TOPMed |
|
|
rs1397431974 CA410765990 |
270 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1171503635 CA410766034 |
272 | N>K | No |
ClinGen gnomAD |
|
|
rs1375676692 CA410766205 |
282 | R>C | No |
ClinGen gnomAD |
|
|
CA410766211 COSM182541 rs1444326760 |
282 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1305274368 CA410766345 |
289 | T>S | No |
ClinGen gnomAD |
|
|
CA322278343 rs1001709647 |
291 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1034579377 CA322278364 |
293 | I>M | No |
ClinGen Ensembl |
|
|
CA410766425 rs1197627340 |
293 | I>T | No |
ClinGen gnomAD |
|
|
rs774096605 CA10117422 |
296 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA410766467 rs1457428290 |
296 | P>S | No |
ClinGen gnomAD |
|
|
rs568979801 CA10117423 |
297 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
rs766986280 CA10117425 |
299 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA10117424 rs766986280 |
299 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA410766502 rs766986280 |
299 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA10117428 rs201830706 |
301 | E>D | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1004094172 CA322278385 |
301 | E>G | No |
ClinGen TOPMed gnomAD |
No associated diseases with P46109
5 regional properties for P46109
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 12 - 102 | IPR000980 |
| domain | SH3 domain | 123 - 183 | IPR001452-1 |
| domain | SH3 domain | 235 - 296 | IPR001452-2 |
| domain | CRK, N-terminal SH3 domain | 126 - 180 | IPR035457 |
| domain | CRK, C-terminal SH3 domain | 237 - 293 | IPR035458 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extrinsic component of postsynaptic membrane | The component of the postsynaptic membrane consisting of gene products and protein complexes that are loosely bound to one of its surfaces, but not integrated into the hydrophobic region. |
| neuromuscular junction | The junction between the axon of a motor neuron and a muscle fiber. In response to the arrival of action potentials, the presynaptic button releases molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane of the muscle fiber, leading to a change in post-synaptic potential. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| identical protein binding | Binding to an identical protein or proteins. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA polymerase II-specific DNA-binding transcription factor binding | Binding to a sequence-specific DNA binding RNA polymerase II transcription factor, any of the factors that interact selectively and non-covalently with a specific DNA sequence in order to modulate transcription. |
| signaling adaptor activity | The binding activity of a molecule that brings together two or more molecules in a signaling pathway, permitting those molecules to function in a coordinated way. Adaptor molecules themselves do not have catalytic activity. |
49 GO annotations of biological process
| Name | Definition |
|---|---|
| acetylcholine receptor signaling pathway | The series of molecular signals generated as a consequence of an acetylcholine receptor binding to one of its physiological ligands. |
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| B cell apoptotic process | Any apoptotic process in a B cell, a lymphocyte of B lineage with the phenotype CD19-positive and capable of B cell mediated immunity. |
| blood vessel development | The process whose specific outcome is the progression of a blood vessel over time, from its formation to the mature structure. The blood vessel is the vasculature carrying blood. |
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cellular response to interleukin-7 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-7 stimulus. |
| cellular response to transforming growth factor beta stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a transforming growth factor beta stimulus. |
| cellular response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organism exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| cerebellar neuron development | The process whose specific outcome is the progression of a cerebellar neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell. |
| cerebral cortex development | The progression of the cerebral cortex over time from its initial formation until its mature state. The cerebral cortex is the outer layered region of the telencephalon. |
| cranial skeletal system development | The process whose specific outcome is the progression of a cranial skeletal system over time, from its formation to the mature structure. The cranial skeletal system is the skeletal subdivision of the head, and includes the skull (cranium plus mandible), pharyngeal and/or hyoid apparatus. |
| dendrite development | The process whose specific outcome is the progression of the dendrite over time, from its formation to the mature structure. |
| endothelin receptor signaling pathway | A G protein-coupled receptor signaling pathway initiated by endothelin binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| establishment of cell polarity | The specification and formation of anisotropic intracellular organization or cell growth patterns. |
| fibroblast growth factor receptor signaling pathway | The series of molecular signals generated as a consequence of a fibroblast growth factor receptor binding to one of its physiological ligands. |
| helper T cell diapedesis | The passage of a helper T cell between the tight junctions of endothelial cells lining blood vessels, typically the fourth and final step of cellular extravasation. |
| hippocampus development | The progression of the hippocampus over time from its initial formation until its mature state. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinases in the downstream tier to transmit a signal within a cell. |
| lipid metabolic process | The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. |
| male gonad development | The process whose specific outcome is the progression of the male gonad over time, from its formation to the mature structure. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of protein phosphorylation | Any process that stops, prevents or reduces the rate of addition of phosphate groups to amino acids within a protein. |
| neuron migration | The characteristic movement of an immature neuron from germinal zones to specific positions where they will reside as they mature. |
| outflow tract morphogenesis | The process in which the anatomical structures of the outflow tract are generated and organized. The outflow tract is the portion of the heart through which blood flows into the arteries. |
| parathyroid gland development | The process whose specific outcome is the progression of the parathyroid gland over time, from its formation to the mature structure. The parathyroid gland is an organ specialised for secretion of parathyroid hormone. |
| pharynx development | The biological process whose specific outcome is the progression of a pharynx from an initial condition to its mature state. The pharynx is the part of the digestive system immediately posterior to the mouth. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of ERK1 and ERK2 cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| positive regulation of glial cell migration | Any process that activates or increases the frequency, rate or extent of glial cell migration. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| positive regulation of substrate adhesion-dependent cell spreading | Any process that activates or increases the frequency, rate or extent of substrate adhesion-dependent cell spreading. |
| Ras protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Ras superfamily of proteins switching to a GTP-bound active state. |
| reelin-mediated signaling pathway | The series of molecular signals initiated by the binding of reelin (a secreted glycoprotein) to a receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| regulation of cell adhesion mediated by integrin | Any process that modulates the frequency, rate, or extent of cell adhesion mediated by integrin. |
| regulation of cell growth | Any process that modulates the frequency, rate, extent or direction of cell growth. |
| regulation of dendrite development | Any process that modulates the frequency, rate or extent of dendrite development. |
| regulation of skeletal muscle acetylcholine-gated channel clustering | Any process that modulates the frequency, rate or extent of skeletal muscle acetylcholine-gated channel clustering. |
| regulation of T cell migration | Any process that modulates the frequency, rate or extent of T cell migration. |
| retinoic acid receptor signaling pathway | The series of molecular signals generated as a consequence of a retinoic acid receptor binding to one of its physiological ligands. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| single fertilization | The union of male and female gametes to form a zygote. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| synapse assembly | The aggregation, arrangement and bonding together of a set of components to form a synapse. This process ends when the synapse is mature (functional). |
| T cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a T cell. |
| thymus development | The process whose specific outcome is the progression of the thymus over time, from its formation to the mature structure. The thymus is a symmetric bi-lobed organ involved primarily in the differentiation of immature to mature T cells, with unique vascular, nervous, epithelial, and lymphoid cell components. |
| urogenital system development | The process whose specific outcome is the progression of the urogenital system over time, from its formation to the mature structure. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q04929 | CRK | Adapter molecule crk | Gallus gallus (Chicken) | SS |
| Q9XYM0 | Crk | Adapter molecule Crk | Drosophila melanogaster (Fruit fly) | SS |
| O43639 | NCK2 | Cytoplasmic protein NCK2 | Homo sapiens (Human) | EV |
| P16333 | NCK1 | Cytoplasmic protein NCK1 | Homo sapiens (Human) | PR |
| P46108 | CRK | Adapter molecule crk | Homo sapiens (Human) | EV |
| Q64010 | Crk | Adapter molecule crk | Mus musculus (Mouse) | EV SS |
| P47941 | Crkl | Crk-like protein | Mus musculus (Mouse) | SS |
| Q63768 | Crk | Adapter molecule crk | Rattus norvegicus (Rat) | SS |
| Q5U2U2 | Crkl | Crk-like protein | Rattus norvegicus (Rat) | SS |
| Q9NHC3 | ced-2 | Cell death abnormality protein 2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSSARFDSSD | RSAWYMGPVS | RQEAQTRLQG | QRHGMFLVRD | SSTCPGDYVL | SVSENSRVSH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YIINSLPNRR | FKIGDQEFDH | LPALLEFYKI | HYLDTTTLIE | PAPRYPSPPM | GSVSAPNLPT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AEDNLEYVRT | LYDFPGNDAE | DLPFKKGEIL | VIIEKPEEQW | WSARNKDGRV | GMIPVPYVEK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LVRSSPHGKH | GNRNSNSYGI | PEPAHAYAQP | QTTTPLPAVS | GSPGAAITPL | PSTQNGPVFA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KAIQKRVPCA | YDKTALALEV | GDIVKVTRMN | INGQWEGEVN | GRKGLFPFTH | VKIFDPQNPD |
| ENE |