P45985
Gene name |
MAP2K4 (JNKK1, MEK4, MKK4, PRKMK4, SEK1, SERK1, SKK1) |
Protein name |
Dual specificity mitogen-activated protein kinase kinase 4 |
Names |
MAP kinase kinase 4, MAPKK 4, JNK-activating kinase 1, MAPK/ERK kinase 4, MEK 4, SAPK/ERK kinase 1, SEK1, Stress-activated protein kinase kinase 1, SAPK kinase 1, SAPKK-1, SAPKK1, c-Jun N-terminal kinase kinase 1, JNKK |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6416 |
EC number |
2.7.12.2: Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
Protein Class |
DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5-RELATED (PTHR48013) |
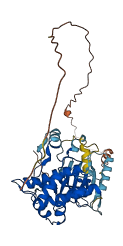
Descriptions
MAP2K4 encodes for Dual specificity mitogen-activated protein kinase kinase 4, belongs to MAP2Ks family, plays an important role in MAP kinase signal transduction pathway. The part of the activation (loop) segment is forming a helical structure (activation segment helix, ASH) in its autoinhibited state, which could be destabilized by the phosphorylation of the activation segment.
Autoinhibitory domains (AIDs)
Accessory elements
246-268 (Activation loop from InterPro)
Target domain |
102-367 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
Structure analysis, Mutagenesis experiment |
References
- Min X et al. (2009) "The structure of the MAP2K MEK6 reveals an autoinhibitory dimer", Structure (London, England : 1993), 17, 96-104
- Shevchenko E et al. (2020) "The autoinhibited state of MKK4: Phosphorylation, putative dimerization and R134W mutant studied by molecular dynamics simulations", Computational and structural biotechnology journal, 18, 2687-2698
Autoinhibited structure

Activated structure
4 structures for P45985
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3ALN | X-ray | 230 A | A/B/C | 80-399 | PDB |
| 3ALO | X-ray | 260 A | A | 80-399 | PDB |
| 3VUT | X-ray | 350 A | A/B | 80-399 | PDB |
| AF-P45985-F1 | Predicted | AlphaFoldDB |
150 variants for P45985
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA398239990 rs1555550018 RCV000505661 |
180 | L>V | Neuroblastoma [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA398214476 rs1417816691 |
4 | P>L | No |
ClinGen TOPMed |
|
|
CA398214482 rs1161471824 |
5 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1161471824 CA398214480 |
5 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
CA288025340 rs979899636 |
6 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs989028622 CA288025362 |
9 | G>D | No |
ClinGen TOPMed |
|
|
CA288025359 rs959911955 |
9 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA288025358 rs959911955 |
9 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA398214514 rs1229178018 |
11 | G>R | No |
ClinGen TOPMed |
|
|
rs749070408 CA8398836 |
15 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA288025374 VAR_062963 rs17855590 |
16 | S>R | No |
ClinGen UniProt TOPMed dbSNP gnomAD |
|
|
CA398214563 rs1169380060 |
18 | S>R | No |
ClinGen TOPMed |
|
|
rs976985059 CA288025382 |
19 | G>D | No |
ClinGen TOPMed |
|
|
rs978668162 CA288025384 |
20 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1289939331 CA398214576 |
21 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA398214582 rs527463657 |
22 | G>C | No |
ClinGen 1000Genomes TOPMed |
|
|
CA288025392 rs527463657 |
22 | G>S | No |
ClinGen 1000Genomes TOPMed |
|
|
rs1283698963 CA398214590 |
23 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs548788415 CA8398838 |
23 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1049103069 CA288025394 |
26 | S>Y | No |
ClinGen TOPMed |
|
|
rs1598004807 CA398214611 |
27 | P>L | No |
ClinGen Ensembl |
|
|
CA288025399 rs910110079 |
28 | A>S | No |
ClinGen TOPMed |
|
|
CA398214626 rs1283304214 |
30 | G>S | No |
ClinGen TOPMed |
|
|
CA398214643 rs1326610085 |
32 | P>Q | No |
ClinGen gnomAD |
|
|
CA398214647 rs1404280869 |
33 | A>T | No |
ClinGen TOPMed |
|
|
CA288025413 rs561204979 |
33 | A>V | No |
ClinGen 1000Genomes gnomAD |
|
|
CA288025420 rs940014479 |
34 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
CA398214651 rs940014479 |
34 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1036999303 CA288025426 |
35 | S>I | No |
ClinGen TOPMed |
|
|
rs1355620714 CA398214681 |
38 | Q>* | No |
ClinGen gnomAD |
|
|
CA398214686 rs1214238261 |
38 | Q>H | No |
ClinGen gnomAD |
|
|
CA8398850 rs760186886 |
41 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs111801039 CA287997640 |
42 | K>R | No |
ClinGen Ensembl |
|
|
rs752767497 CA8398852 |
45 | K>M | No |
ClinGen ExAC gnomAD |
|
|
CA8398853 rs559800950 |
46 | L>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1348623447 CA398203507 |
47 | N>S | No |
ClinGen gnomAD |
|
|
CA287997688 rs1010543298 |
50 | N>S | No |
ClinGen Ensembl |
|
|
rs1447612385 CA398203569 |
50 | N>Y | No |
ClinGen TOPMed |
|
|
rs1261150954 CA398203592 |
51 | P>L | No |
ClinGen TOPMed |
|
|
rs754035296 CA8398855 |
52 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs1358824984 COSM1686247 CA398203612 |
52 | P>L | skin [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs754035296 CA398203604 |
52 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA287997693 rs372872972 |
53 | F>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1348785378 CA398203722 |
57 | A>G | No |
ClinGen gnomAD |
|
|
rs757051936 CA8398856 |
57 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1213129161 CA398203759 |
59 | F>L | No |
ClinGen gnomAD |
|
|
CA8398858 rs750208931 |
63 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA8398857 rs778746854 |
63 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs758381870 CA8398859 |
64 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs780013239 CA8398860 |
66 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201981451 CA287997743 |
68 | V>A | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1394118458 CA398203862 |
69 | Q>E | No |
ClinGen gnomAD |
|
|
rs1434938057 CA398203876 |
70 | N>S | No |
ClinGen gnomAD |
|
|
CA8398886 rs377490722 |
76 | L>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA398208127 rs1290976146 |
90 | S>Y | No |
ClinGen gnomAD |
|
|
rs1466199105 CA398208154 |
94 | H>Y | No |
ClinGen TOPMed |
|
|
CA288017479 rs773665341 |
105 | L>F | No |
ClinGen gnomAD |
|
|
CA398208338 rs1474208027 |
108 | I>F | No |
ClinGen gnomAD |
|
|
CA398208341 rs1215001531 |
108 | I>T | No |
ClinGen TOPMed |
|
|
CA398208364 rs1164096095 COSM79380 |
110 | R>* | ovary endometrium stomach [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1418420168 CA398208374 |
111 | G>* | No |
ClinGen gnomAD |
|
|
CA8398893 rs747647990 |
117 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA398208467 rs1290755523 |
118 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
rs769619151 CA8398894 |
120 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769619151 CA398208496 |
120 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA288017491 rs77267737 |
130 | V>G | No |
ClinGen Ensembl |
|
|
CA398239147 rs1415316526 |
136 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
COSM975559 CA8398968 rs775475276 |
137 | V>M | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1444656785 CA398239163 |
138 | D>N | No |
ClinGen TOPMed |
|
|
rs1371329502 CA398239180 |
139 | E>K | No |
ClinGen TOPMed |
|
|
CA398239277 rs1307654543 |
146 | L>V | No |
ClinGen TOPMed |
|
|
rs760515884 CA8398969 |
147 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8398971 rs776387940 |
151 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA8398972 rs761402309 |
153 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA398239334 rs1208900089 |
154 | R>Q | No |
ClinGen gnomAD |
|
|
CA398239332 rs1567657403 VAR_040819 COSM20411 |
154 | R>W | large_intestine a colorectal adenocarcinoma sample; somatic mutation [Cosmic, UniProt] | No |
ClinGen cosmic curated UniProt Ensembl dbSNP |
|
rs1314005490 CA398239360 |
157 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs764582118 CA8398973 |
159 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs1248970628 CA398239377 |
160 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs749942107 CA8398974 |
160 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA398239412 rs1418890158 |
165 | Y>N | No |
ClinGen gnomAD |
|
|
rs534883661 CA287984272 |
178 | M>V | No |
ClinGen Ensembl |
|
|
rs1191386417 CA398240022 |
182 | S>C | No |
ClinGen gnomAD |
|
|
rs766050183 CA8398995 |
185 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA8398996 rs150905259 |
191 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs760844487 CA287984326 |
195 | V>A | No |
ClinGen Ensembl |
|
|
rs764298113 CA8398998 |
195 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs754217305 CA8398999 |
196 | L>S | No |
ClinGen ExAC gnomAD |
|
|
CA398240251 rs1371909082 |
200 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1040630581 CA287984336 |
203 | E>D | No |
ClinGen TOPMed |
|
|
CA8399001 rs765746113 |
207 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1307489818 CA398240352 |
208 | I>T | No |
ClinGen gnomAD |
|
|
CA398240362 rs1260446708 |
209 | T>S | No |
ClinGen gnomAD |
|
|
CA398240370 rs1240723113 |
210 | L>S | No |
ClinGen gnomAD |
|
|
rs1354928111 CA398241122 |
222 | N>S | No |
ClinGen TOPMed |
|
|
CA287988423 rs748395840 |
230 | I>T | No |
ClinGen gnomAD |
|
|
CA398241369 rs1597480176 |
231 | K>N | No |
ClinGen Ensembl |
|
|
rs536223681 CA8399033 |
231 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1025397785 CA287988437 |
242 | N>S | No |
ClinGen TOPMed |
|
|
CA398241507 rs1160578914 |
243 | I>T | No |
ClinGen gnomAD |
|
|
rs1361335563 CA398241605 |
255 | V>L | No |
ClinGen gnomAD |
|
|
CA287988456 rs200798818 |
264 | A>S | No |
ClinGen 1000Genomes |
|
|
rs76726054 CA287988466 |
266 | C>W | No |
ClinGen Ensembl |
|
|
rs1455763623 CA398241772 |
277 | P>A | No |
ClinGen TOPMed |
|
|
COSM21292 VAR_040822 CA8399062 rs753665559 |
279 | A>T | large_intestine haematopoietic_and_lymphoid_tissue a colorectal adenocarcinoma sample; somatic mutation [Cosmic, UniProt] | No |
ClinGen cosmic curated UniProt ExAC dbSNP gnomAD |
|
rs1183392154 COSM1563407 CA398241789 |
279 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA8399063 rs537815982 |
286 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA398241845 rs1334746981 |
288 | S>C | No |
ClinGen gnomAD |
|
|
CA8399090 rs144008624 |
301 | A>G | No |
ClinGen ESP ExAC |
|
|
rs781002756 CA8399089 |
301 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1194360851 CA398241966 |
304 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs755823478 CA8399091 |
309 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs779390733 CA8399092 |
311 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs1480785686 CA398242019 |
311 | N>K | No |
ClinGen TOPMed |
|
|
rs1396947681 COSM98424 CA398242084 |
321 | V>M | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA288003875 COSM98425 rs267604739 |
326 | P>L | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1311024915 CA398242129 |
327 | Q>H | No |
ClinGen gnomAD |
|
|
CA398242154 rs1597497681 |
331 | S>Y | No |
ClinGen Ensembl |
|
|
CA398242159 rs1355543573 |
332 | E>K | No |
ClinGen gnomAD |
|
|
rs908877597 CA288003883 |
335 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA8399100 rs371746849 |
338 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA398242205 rs371746849 |
338 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA288003902 rs367710581 |
341 | I>T | No |
ClinGen ESP TOPMed |
|
|
rs1239669542 CA398242223 |
341 | I>V | No |
ClinGen gnomAD |
|
|
rs1355990355 CA398242234 |
342 | N>K | No |
ClinGen gnomAD |
|
|
CA8399102 rs191244765 |
342 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs755523599 CA8399132 COSM975566 |
349 | T>M | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1293062852 CA398242297 |
349 | T>S | No |
ClinGen gnomAD |
|
|
rs1294064382 CA398242316 |
351 | D>E | No |
ClinGen gnomAD |
|
|
rs1340390247 CA398242321 |
352 | E>G | No |
ClinGen gnomAD |
|
|
CA288014390 rs1024493228 |
355 | R>M | No |
ClinGen TOPMed |
|
|
CA8399135 rs769700685 |
359 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA398242410 rs1567679461 |
363 | K>R | No |
ClinGen Ensembl |
|
|
CA398242415 rs1323806098 |
364 | H>Y | No |
ClinGen TOPMed |
|
|
CA398242443 rs1425777448 |
368 | L>V | No |
ClinGen gnomAD |
|
|
CA398242461 rs1379788876 |
370 | Y>C | No |
ClinGen gnomAD |
|
|
rs749364956 CA8399156 |
370 | Y>N | No |
ClinGen ExAC gnomAD |
|
|
CA398242465 rs1168805084 |
371 | E>K | No |
ClinGen gnomAD |
|
|
rs1300593472 CA398242483 |
373 | R>C | No |
ClinGen gnomAD |
|
|
rs1372846289 CA398242484 |
373 | R>H | No |
ClinGen gnomAD |
|
|
CA398242487 rs1052408848 |
374 | A>S | No |
ClinGen TOPMed |
|
|
CA288015641 rs1052408848 |
374 | A>T | No |
ClinGen TOPMed |
|
|
rs889363169 CA288015651 |
374 | A>V | No |
ClinGen TOPMed |
|
|
rs367662793 CA8399158 |
375 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs771926980 CA8399160 |
378 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs771926980 CA8399161 COSM975568 |
378 | A>T | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA398242567 rs1192323091 |
386 | D>A | No |
ClinGen TOPMed |
|
|
CA398242633 rs1209244324 |
396 | M>V | No |
ClinGen gnomAD |
|
|
rs538492504 CA8399165 |
397 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1177038824 CA398242655 |
399 | D>N | No |
ClinGen gnomAD |
No associated diseases with P45985
5 regional properties for P45985
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Formin, FH3 domain | 284 - 637 | IPR010472 |
| domain | Formin, GTPase-binding domain | 27 - 281 | IPR010473 |
| domain | Diaphanous autoregulatory (DAD) domain | 1059 - 1090 | IPR014767 |
| domain | Rho GTPase-binding/formin homology 3 (GBD/FH3) domain | 27 - 468 | IPR014768 |
| domain | Formin, FH2 domain | 632 - 1067 | IPR015425 |
Functions
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| JUN kinase kinase activity | Catalysis of the phosphorylation of tyrosine and threonine residues in a c-Jun NH2-terminal kinase (JNK), a member of a subgroup of mitogen-activated protein kinases (MAPKs), which signal in response to cytokines and exposure to environmental stress. JUN kinase kinase (JNKK) is a dual-specificity protein kinase kinase and requires activation by a serine/threonine kinase JUN kinase kinase kinase. |
| MAP kinase kinase activity | Catalysis of the concomitant phosphorylation of threonine (T) and tyrosine (Y) residues in a Thr-Glu-Tyr (TEY) thiolester sequence in a MAP kinase (MAPK) substrate. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cellular response to mechanical stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| cellular senescence | A cell aging process stimulated in response to cellular stress, whereby normal cells lose the ability to divide through irreversible cell cycle arrest. |
| Fc-epsilon receptor signaling pathway | The series of molecular signals initiated by the binding of the Fc portion of immunoglobulin E (IgE) to an Fc-epsilon receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. The Fc portion of an immunoglobulin is its C-terminal constant region. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinases in the downstream tier to transmit a signal within a cell. |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinase in the downstream tier to transmit a signal within a cell. |
| negative regulation of motor neuron apoptotic process | Any process that stops, prevents or reduces the frequency, rate or extent of motor neuron apoptotic process. |
| positive regulation of smooth muscle cell apoptotic process | Any process that activates or increases the frequency, rate, or extent of smooth muscle cell apoptotic process. |
| response to wounding | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9X2 | MAP2K6 | Dual specificity mitogen-activated protein kinase kinase 6 | Bos taurus (Bovine) | EV |
| O14733 | MAP2K7 | Dual specificity mitogen-activated protein kinase kinase 7 | Homo sapiens (Human) | PR |
| P52564 | MAP2K6 | Dual specificity mitogen-activated protein kinase kinase 6 | Homo sapiens (Human) | EV |
| P46734 | MAP2K3 | Dual specificity mitogen-activated protein kinase kinase 3 | Homo sapiens (Human) | SS |
| Q13163 | MAP2K5 | Dual specificity mitogen-activated protein kinase kinase 5 | Homo sapiens (Human) | PR |
| P36507 | MAP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | Homo sapiens (Human) | EV |
| Q02750 | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 | Homo sapiens (Human) | EV |
| P47809 | Map2k4 | Dual specificity mitogen-activated protein kinase kinase 4 | Mus musculus (Mouse) | EV |
| P70236 | Map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Mus musculus (Mouse) | EV |
| Q8CE90 | Map2k7 | Dual specificity mitogen-activated protein kinase kinase 7 | Mus musculus (Mouse) | PR |
| O09110 | Map2k3 | Dual specificity mitogen-activated protein kinase kinase 3 | Mus musculus (Mouse) | PR |
| G5EDF7 | sek-1 | Dual specificity mitogen-activated protein kinase kinase sek-1 | Caenorhabditis elegans | PR |
| G5EDT6 | jkk-1 | Dual specificity mitogen-activated protein kinase kinase jkk-1 | Caenorhabditis elegans | PR |
| Q21307 | mek-1 | Dual specificity mitogen-activated protein kinase kinase mek-1 | Caenorhabditis elegans | PR |
| Q9DGE0 | map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAPSPSGGG | GSGGGSGSGT | PGPVGSPAPG | HPAVSSMQGK | RKALKLNFAN | PPFKSTARFT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LNPNPTGVQN | PHIERLRTHS | IESSGKLKIS | PEQHWDFTAE | DLKDLGEIGR | GAYGSVNKMV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| HKPSGQIMAV | KRIRSTVDEK | EQKQLLMDLD | VVMRSSDCPY | IVQFYGALFR | EGDCWICMEL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MSTSFDKFYK | YVYSVLDDVI | PEEILGKITL | ATVKALNHLK | ENLKIIHRDI | KPSNILLDRS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GNIKLCDFGI | SGQLVDSIAK | TRDAGCRPYM | APERIDPSAS | RQGYDVRSDV | WSLGITLYEL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ATGRFPYPKW | NSVFDQLTQV | VKGDPPQLSN | SEEREFSPSF | INFVNLCLTK | DESKRPKYKE |
| 370 | 380 | 390 | |||
| LLKHPFILMY | EERAVEVACY | VCKILDQMPA | TPSSPMYVD |