P45983
Gene name |
MAPK8 (JNK1, PRKM8, SAPK1, SAPK1C) |
Protein name |
Mitogen-activated protein kinase 8 |
Names |
MAP kinase 8 , MAPK 8 , EC 2.7.11.24 , JNK-46 , Stress-activated protein kinase 1c , SAPK1c , Stress-activated protein kinase JNK1 , c-Jun N-terminal kinase 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5599 |
EC number |
2.7.11.24: Protein-serine/threonine kinases |
Protein Class |
MITOGEN-ACTIVATED PROTEIN KINASE (PTHR24055) |
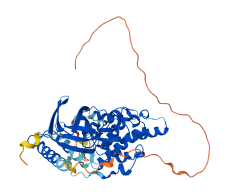
Descriptions
JNK1 is a member of the mitogen-activated protein kinase (MAPK) family, involved in various processes such as cell proliferation, differentiation, migration, transformation and programmed cell death. It undergoes autoinhibition through peptide-induced interlobe opening and rotation, moving αC out of the catalytic site and allowing the activation loop to form an inhibitory helix blocking the ATP binding site of JNK1’s kinase domain. Autoinhibition is relieved by phosphorylation of the activation loop.
Autoinhibitory domains (AIDs)
Accessory elements
168-178 (Activation loop from InterPro)
Target domain |
26-321 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis, Mutagenesis experiment |
182-190 (Activation loop from InterPro)
Target domain |
26-321 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis, Mutagenesis experiment |
Autoinhibited structure

Activated structure

35 structures for P45983
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1UKH | X-ray | 235 A | A | 1-363 | PDB |
| 1UKI | X-ray | 270 A | A | 1-363 | PDB |
| 2G01 | X-ray | 350 A | A/B | 1-364 | PDB |
| 2GMX | X-ray | 350 A | A/B | 1-364 | PDB |
| 2H96 | X-ray | 300 A | A/B | 1-364 | PDB |
| 2NO3 | X-ray | 320 A | A/B | 1-364 | PDB |
| 2XRW | X-ray | 133 A | A | 2-364 | PDB |
| 2XS0 | X-ray | 260 A | A | 1-379 | PDB |
| 3ELJ | X-ray | 180 A | A | 1-364 | PDB |
| 3O17 | X-ray | 300 A | A/B | 1-364 | PDB |
| 3O2M | X-ray | 270 A | A/B | 1-364 | PDB |
| 3PZE | X-ray | 200 A | A | 7-364 | PDB |
| 3V3V | X-ray | 270 A | A | 1-366 | PDB |
| 3VUD | X-ray | 350 A | A | 1-364 | PDB |
| 3VUG | X-ray | 324 A | A | 1-364 | PDB |
| 3VUH | X-ray | 270 A | A | 1-364 | PDB |
| 3VUI | X-ray | 280 A | A | 1-364 | PDB |
| 3VUK | X-ray | 295 A | A | 1-364 | PDB |
| 3VUL | X-ray | 281 A | A | 1-364 | PDB |
| 3VUM | X-ray | 269 A | A | 1-364 | PDB |
| 4AWI | X-ray | 191 A | A | 1-364 | PDB |
| 4E73 | X-ray | 227 A | A | 1-363 | PDB |
| 4G1W | X-ray | 245 A | A | 1-363 | PDB |
| 4HYS | X-ray | 242 A | A | 1-363 | PDB |
| 4HYU | X-ray | 215 A | A | 1-363 | PDB |
| 4IZY | X-ray | 230 A | A | 1-363 | PDB |
| 4L7F | X-ray | 195 A | A | 7-362 | PDB |
| 4QTD | X-ray | 150 A | A | 1-363 | PDB |
| 4UX9 | X-ray | 234 A | A/B/C/D | 1-364 | PDB |
| 4YR8 | X-ray | 240 A | A/C/E/F | 1-363 | PDB |
| 5LW1 | X-ray | 320 A | B/E/H | 2-363 | PDB |
| 6F5E | X-ray | 270 A | B | 2-363 | PDB |
| 6ZR5 | X-ray | 270 A | A/B | 1-364 | PDB |
| 8X5M | X-ray | 200 A | A | 2-364 | PDB |
| AF-P45983-F1 | Predicted | AlphaFoldDB |
290 variants for P45983
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs770396528 | 2 | S>N | No |
ExAC gnomAD |
|
| rs2042165911 | 3 | R>G | No | Ensembl | |
| rs2042166036 | 4 | S>C | No | Ensembl | |
| rs2042166193 | 4 | S>R | No | Ensembl | |
|
COSM5074514 COSM170014 rs773780784 |
6 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
COSM4014379 COSM4014380 rs759178320 COSM4014381 |
6 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs773780784 | 6 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2042166874 | 7 | D>G | No | TOPMed | |
| rs771655862 | 8 | N>S | No |
ExAC gnomAD |
|
| rs1467885822 | 9 | N>D | No | gnomAD | |
| rs150753150 | 9 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1047240487 | 9 | N>S | No |
TOPMed gnomAD |
|
| rs1406003607 | 10 | F>I | No |
TOPMed gnomAD |
|
| rs1299206807 | 11 | Y>C | No | gnomAD | |
| rs760960935 | 11 | Y>H | No |
ExAC gnomAD |
|
| rs754396010 | 14 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs764612003 | 14 | E>K | No |
ExAC gnomAD |
|
| rs2042169022 | 17 | D>E | No | Ensembl | |
| rs1233570131 | 17 | D>V | No |
TOPMed gnomAD |
|
| rs765388602 | 19 | T>A | No |
ExAC gnomAD |
|
| rs370411166 | 22 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2132977018 | 23 | L>M | No | Ensembl | |
| rs780150000 | 24 | K>T | No |
ExAC TOPMed gnomAD |
|
|
COSM427595 rs2042170101 COSM5196829 COSM427596 |
25 | R>* | Variant assessed as Somatic; HIGH impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
|
COSM5349138 COSM5349139 COSM216920 rs755894470 |
25 | R>Q | liver Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1318665915 | 26 | Y>* | No |
TOPMed gnomAD |
|
| rs2042170781 | 28 | N>K | No | Ensembl | |
|
COSM918463 COSM918462 rs1417658483 |
31 | P>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs2132977453 | 32 | I>T | No | Ensembl | |
| rs1481506037 | 32 | I>V | No |
TOPMed gnomAD |
|
|
COSM1347947 COSM1347948 COSM5135354 |
38 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1664423061 | 39 | I>T | No |
TOPMed gnomAD |
|
| rs1323103105 | 39 | I>V | No | gnomAD | |
|
COSM918464 COSM918465 rs778730942 COSM5165497 |
41 | C>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs770874861 | 41 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs200350498 | 42 | A>S | No |
ExAC TOPMed gnomAD |
|
|
rs200350498 COSM1347950 COSM1347949 COSM4602711 |
42 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1361641151 | 43 | A>G | No | gnomAD | |
| rs1589210960 | 43 | A>T | No | Ensembl | |
| rs2042377460 | 44 | Y>F | No | Ensembl | |
| rs1467645843 | 44 | Y>H | No | gnomAD | |
| rs143875999 | 46 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs779657933 | 46 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2042377938 | 47 | I>V | No | Ensembl | |
| rs746642373 | 48 | L>P | No |
ExAC gnomAD |
|
| rs1233762287 | 49 | E>G | No | gnomAD | |
| rs2042378605 | 51 | N>H | No | Ensembl | |
| rs2042378763 | 51 | N>S | No | TOPMed | |
| rs2042379221 | 53 | A>G | No | gnomAD | |
| rs2042379049 | 53 | A>T | No | TOPMed | |
| rs2133024036 | 54 | I>V | No | Ensembl | |
| rs1157307365 | 58 | S>G | No | gnomAD | |
| rs554758335 | 59 | R>* | No |
1000Genomes ExAC gnomAD |
|
| rs573076590 | 59 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1363332287 | 68 | K>N | No | gnomAD | |
| rs1411261165 | 69 | R>Q | No | gnomAD | |
|
COSM918467 COSM918466 |
73 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1739252749 | 76 | L>F | No | gnomAD | |
|
COSM1347954 COSM5171750 COSM1347953 |
77 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1334928589 | 77 | M>T | No | gnomAD | |
| rs2042381603 | 79 | C>S | No | gnomAD | |
| rs2133024708 | 80 | V>A | No | Ensembl | |
| rs1439986144 | 80 | V>I | No | gnomAD | |
| rs2042381911 | 83 | K>N | No | gnomAD | |
| TCGA novel | 84 | N>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1029787979 | 85 | I>V | No | TOPMed | |
| rs780976087 | 90 | N>H | No |
ExAC gnomAD |
|
| rs200881923 | 94 | P>R | No | 1000Genomes | |
| TCGA novel | 95 | Q>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773674759 | 97 | S>C | No |
ExAC TOPMed gnomAD |
|
|
COSM3438211 COSM1702325 rs773674759 COSM1702324 |
97 | S>F | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2042660731 | 98 | L>P | No | TOPMed | |
|
COSM918469 rs2042661088 COSM918470 |
101 | F>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
|
COSM190535 COSM918468 |
101 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771498910 | 102 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 103 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1444972184 | 105 | Y>* | No | gnomAD | |
| rs750668630 | 106 | I>M | No | Ensembl | |
| rs2042667596 | 106 | I>V | No | gnomAD | |
| rs752372488 | 110 | L>R | No |
ExAC gnomAD |
|
| rs2042668485 | 112 | D>G | No | Ensembl | |
| rs755812386 | 113 | A>G | No |
ExAC gnomAD |
|
| rs2042668649 | 113 | A>T | No |
TOPMed gnomAD |
|
|
COSM5560673 COSM5560674 COSM5560675 |
114 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM257372 COSM918471 |
115 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3438213 COSM3438212 COSM3438214 |
121 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1173910920 | 121 | M>V | No | gnomAD | |
| rs1418848027 | 122 | E>A | No |
TOPMed gnomAD |
|
| rs1418848027 | 122 | E>V | No |
TOPMed gnomAD |
|
|
COSM3967075 COSM3967076 COSM3967077 |
125 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1181281990 | 128 | M>I | No |
TOPMed gnomAD |
|
| rs2042670235 | 131 | L>R | No | TOPMed | |
| rs1196726534 | 135 | M>L | No |
TOPMed gnomAD |
|
| rs1196726534 | 135 | M>V | No |
TOPMed gnomAD |
|
| rs1215027038 | 143 | H>N | No |
TOPMed gnomAD |
|
| rs779380684 | 145 | A>V | No |
ExAC gnomAD |
|
| rs1398431451 | 146 | G>V | No | gnomAD | |
| rs2042672007 | 148 | I>V | No | gnomAD | |
|
COSM684193 COSM684192 |
149 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs755655560 | 155 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs2043275782 | 155 | S>R | No | Ensembl | |
| rs1451383737 | 157 | I>M | No | TOPMed | |
| rs368198154 | 157 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1220824711 | 163 | C>F | No |
TOPMed gnomAD |
|
| TCGA novel | 166 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_042258 | 171 | G>S | a renal clear cell carcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs2043277231 | 172 | L>P | No | Ensembl | |
| rs868234992 | 173 | A>V | No | Ensembl | |
| rs372126348 | 176 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM13337 rs2043278229 VAR_042259 |
177 | G>R | central_nervous_system a glioblastoma multiforme sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated gnomAD UniProt |
|
COSM4827338 COSM4827337 rs1162940624 COSM4827336 |
178 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1356700893 | 182 | M>V | No | gnomAD | |
|
COSM918472 COSM918473 |
183 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2043279063 | 186 | V>E | No | TOPMed | |
| TCGA novel | 188 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM684190 rs202013888 COSM684191 |
189 | R>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
|
COSM918474 COSM918475 |
189 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM684186 COSM684187 |
189 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1589253309 | 194 | P>S | No | Ensembl | |
| rs868332724 | 198 | L>P | No | Ensembl | |
| rs1037196002 | 205 | N>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 206 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2043526046 | 209 | W>G | No |
TOPMed gnomAD |
|
| rs1199564380 | 211 | V>L | No | TOPMed | |
| rs747531271 | 214 | I>S | No |
ExAC gnomAD |
|
| rs2043527558 | 215 | M>V | No | Ensembl | |
| rs1262446655 | 216 | G>E | No | gnomAD | |
| COSM3415062 | 217 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM918476 | 217 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2043527957 | 218 | M>T | No |
TOPMed gnomAD |
|
| TCGA novel | 221 | H>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2043528776 | 226 | P>A | No | Ensembl | |
| rs376010774 | 226 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs774221220 | 229 | D>N | No |
ExAC gnomAD |
|
| rs1402727389 | 231 | I>L | No |
TOPMed gnomAD |
|
|
rs1402727389 COSM190546 |
231 | I>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA TOPMed gnomAD |
| rs748025349 | 235 | N>S | No |
ExAC gnomAD |
|
| rs2043653013 | 240 | Q>H | No | TOPMed | |
|
COSM3790810 COSM3790809 |
240 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 241 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1297286 COSM1297287 |
246 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM415015 COSM415014 |
247 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756073891 | 249 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs377156368 | 249 | M>T | No |
ESP TOPMed gnomAD |
|
| rs756073891 | 249 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs2043654040 | 251 | K>R | No | Ensembl | |
| rs780732703 | 253 | Q>* | No | Ensembl | |
| rs2043654875 | 256 | V>I | No | Ensembl | |
| rs1589269014 | 258 | T>A | No | Ensembl | |
| rs1352216003 | 260 | V>A | No | gnomAD | |
|
COSM1347970 COSM1347971 |
260 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1205418935 | 262 | N>S | No | gnomAD | |
| rs770530313 | 263 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs752183776 | 264 | P>T | No | Ensembl | |
| rs1201342024 | 266 | Y>F | No | gnomAD | |
|
COSM3438218 COSM3438219 |
266 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1269301566 | 267 | A>S | No |
TOPMed gnomAD |
|
| rs1269301566 | 267 | A>T | No |
TOPMed gnomAD |
|
| rs2043657678 | 268 | G>R | No | TOPMed | |
| rs2043658283 | 269 | Y>C | No | gnomAD | |
| rs199509878 | 269 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| rs758356952 | 270 | S>C | No | Ensembl | |
| rs569412118 | 272 | E>A | No | 1000Genomes | |
| TCGA novel | 275 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4014383 COSM4014384 |
276 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 280 | F>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM415012 COSM415013 |
283 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1290318874 | 283 | D>N | No | TOPMed | |
| rs2043660231 | 287 | N>D | No | Ensembl | |
| rs1430973969 | 288 | K>Q | No | Ensembl | |
| rs2043660607 | 288 | K>R | No | Ensembl | |
| rs1159852187 | 292 | S>G | No |
TOPMed gnomAD |
|
| rs1448138314 | 292 | S>N | No | gnomAD | |
| rs1454207254 | 293 | Q>R | No |
TOPMed gnomAD |
|
| rs2043686720 | 297 | L>V | No |
TOPMed gnomAD |
|
| rs2043686904 | 299 | S>P | No | Ensembl | |
| rs2043687080 | 301 | M>L | No | Ensembl | |
| rs2043687877 | 304 | I>L | No | TOPMed | |
| rs45505100 | 304 | I>M | No |
ExAC gnomAD |
|
| rs1313003666 | 306 | A>E | No | gnomAD | |
| rs775931291 | 306 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1320670600 | 308 | K>E | No | gnomAD | |
| rs1242378993 | 308 | K>R | No | gnomAD | |
| rs747392955 | 311 | S>C | No |
ExAC gnomAD |
|
| rs540242907 | 313 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1237716763 | 313 | D>G | No |
TOPMed gnomAD |
|
| rs769203778 | 313 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs769203778 | 313 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1185435366 | 314 | E>K | No | gnomAD | |
| TCGA novel | 316 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 320 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs142342900 | 322 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 325 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1454123136 | 330 | A>T | No | gnomAD | |
| rs2043692546 | 332 | A>G | No | Ensembl | |
| rs986626073 | 334 | P>L | No | gnomAD | |
| rs763747268 | 336 | K>R | No |
ExAC gnomAD |
|
| rs1427342522 | 337 | I>M | No | Ensembl | |
| rs2043731220 | 338 | P>H | No | TOPMed | |
| rs1276026393 | 338 | P>S | No |
TOPMed gnomAD |
|
| rs1276026393 | 338 | P>T | No |
TOPMed gnomAD |
|
| rs2043731418 | 339 | D>H | No | TOPMed | |
|
COSM4014386 COSM4014385 |
339 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1475171749 | 340 | K>N | No |
TOPMed gnomAD |
|
|
COSM1347973 COSM1347972 |
341 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2043731777 | 343 | D>N | No | TOPMed | |
| rs753698159 | 345 | R>K | No |
ExAC gnomAD |
|
| rs765234973 | 346 | E>K | No |
ExAC gnomAD |
|
| rs750388057 | 347 | H>Y | No |
ExAC gnomAD |
|
| rs757980939 | 348 | T>A | No |
ExAC gnomAD |
|
| rs1390438534 | 349 | I>T | No |
TOPMed gnomAD |
|
| rs1488717032 | 349 | I>V | No |
TOPMed gnomAD |
|
| rs1328803228 | 350 | E>D | No |
TOPMed gnomAD |
|
| rs944850013 | 353 | K>R | No |
TOPMed gnomAD |
|
| rs1354748570 | 355 | L>M | No |
TOPMed gnomAD |
|
| rs2044217039 | 356 | I>M | No | Ensembl | |
| rs754818843 | 357 | Y>C | No |
TOPMed gnomAD |
|
| rs201417282 | 357 | Y>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs371493413 | 361 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs45483593 VAR_050592 |
365 | E>K | No |
UniProt Ensembl dbSNP |
|
| rs2044219561 | 366 | R>G | No | TOPMed | |
| rs28395809 | 367 | T>A | No | Ensembl | |
| rs751150249 | 367 | T>S | No |
ExAC gnomAD |
|
| rs1346038846 | 368 | K>E | No | gnomAD | |
| rs767178901 | 372 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs988262121 | 372 | I>T | No | gnomAD | |
| rs767178901 | 372 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs756498386 | 373 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs756498386 | 373 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs752334346 | 373 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs151285118 | 374 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs151285118 | 374 | G>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs778058488 | 374 | G>R | No |
ExAC gnomAD |
|
| rs1348073540 | 376 | P>L | No | gnomAD | |
| rs1348073540 | 376 | P>R | No | gnomAD | |
| rs778924369 | 377 | S>A | No |
ExAC gnomAD |
|
| rs778924369 | 377 | S>P | No |
ExAC gnomAD |
|
| rs745968638 | 378 | P>R | No |
ExAC gnomAD |
|
| rs2133327919 | 378 | P>S | No | Ensembl | |
| rs1019024052 | 380 | G>V | No | Ensembl | |
| rs1323743097 | 382 | A>T | No | TOPMed | |
| rs2044726922 | 384 | I>M | No | Ensembl | |
| rs758497860 | 384 | I>N | No |
ExAC gnomAD |
|
| rs780052742 | 385 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs780052742 | 385 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs747193295 | 387 | S>F | No |
ExAC gnomAD |
|
| rs768761581 | 389 | H>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 389 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749073014 | 392 | S>A | No |
ExAC gnomAD |
|
| rs1422869920 | 393 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2044729666 | 394 | S>L | No | TOPMed | |
| rs933003988 | 395 | S>C | No |
TOPMed gnomAD |
|
| TCGA novel | 395 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759451457 | 397 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs367599489 | 397 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2044731322 | 398 | D>Y | No | TOPMed | |
| rs1353720299 | 399 | V>A | No |
TOPMed gnomAD |
|
| rs992324943 | 399 | V>M | No | gnomAD | |
| rs763758430 | 402 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs774869055 | 402 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs774869055 | 402 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1285833471 | 404 | T>A | No | Ensembl | |
| rs1340500693 | 404 | T>I | No | gnomAD | |
| TCGA novel | 405 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765655352 | 406 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1199331193 | 408 | L>S | No | gnomAD | |
| rs2044734894 | 409 | A>D | No | Ensembl | |
| rs1276075262 | 410 | S>C | No |
TOPMed gnomAD |
|
| rs1276075262 | 410 | S>F | No |
TOPMed gnomAD |
|
| rs2044735690 | 412 | T>K | No | Ensembl | |
| rs2044735940 | 414 | S>R | No | TOPMed | |
| rs1489044523 | 416 | L>R | No | gnomAD | |
| rs147673874 | 418 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs147673874 | 418 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs972332361 | 419 | A>S | No | TOPMed | |
| rs972332361 | 419 | A>T | No | TOPMed | |
| rs142606131 | 420 | A>D | No |
ESP ExAC gnomAD |
|
| rs1011736095 | 420 | A>P | No | gnomAD | |
| rs2044738415 | 421 | G>R | No | Ensembl | |
| rs1589341428 | 423 | L>Q | No | Ensembl | |
| rs1589341428 | 423 | L>R | No | Ensembl | |
| rs1325673457 | 423 | L>V | No | gnomAD | |
| rs781538114 | 424 | G>D | No |
ExAC gnomAD |
|
| rs781538114 | 424 | G>V | No |
ExAC gnomAD |
|
| rs2044740086 | 425 | C>F | No | Ensembl | |
| rs2044739854 | 425 | C>G | No | Ensembl | |
| rs748451475 | 427 | R>G | No |
ExAC gnomAD |
|
| rs1478396994 | 427 | R>I | No | gnomAD | |
| rs2044740784 | 427 | R>S | No | Ensembl | |
| rs2044741039 | 428 | R>R | No | TOPMed |
No associated diseases with P45983
Functions
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| basal dendrite | A dendrite that emerges near the basal pole of a neuron. In bipolar neurons, basal dendrites are either on the same side of the soma as the axon, or project toward the axon. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| histone deacetylase binding | Binding to histone deacetylase. |
| histone deacetylase regulator activity | Binds to and modulates the activity of histone deacetylase. |
| JUN kinase activity | Catalysis of the reaction |
| protein phosphatase binding | Binding to a protein phosphatase. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase binding | Binding to a protein serine/threonine kinase. |
30 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to amino acid starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of amino acids. |
| cellular response to cadmium ion | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cadmium (Cd) ion stimulus. |
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| cellular response to mechanical stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| cellular response to reactive oxygen species | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a reactive oxygen species stimulus. Reactive oxygen species include singlet oxygen, superoxide, and oxygen free radicals. |
| cellular senescence | A cell aging process stimulated in response to cellular stress, whereby normal cells lose the ability to divide through irreversible cell cycle arrest. |
| Fc-epsilon receptor signaling pathway | The series of molecular signals initiated by the binding of the Fc portion of immunoglobulin E (IgE) to an Fc-epsilon receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. The Fc portion of an immunoglobulin is its C-terminal constant region. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier |
| JUN phosphorylation | The process of introducing a phosphate group into a JUN protein. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of protein binding | Any process that stops, prevents, or reduces the frequency, rate or extent of protein binding. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peptidyl-threonine phosphorylation | The phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of cell killing | Any process that activates or increases the frequency, rate or extent of cell killing. |
| positive regulation of cyclase activity | Any process that activates or increases the activity of a cyclase. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of NLRP3 inflammasome complex assembly | Any process that activates or increases the frequency, rate or extent of NLRP3 inflammasome complex assembly. |
| positive regulation of protein metabolic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of circadian rhythm | Any process that modulates the frequency, rate or extent of a circadian rhythm. A circadian rhythm is a biological process in an organism that recurs with a regularity of approximately 24 hours. |
| regulation of DNA replication origin binding | Any process that modulates the frequency, rate or extent of DNA replication origin binding. |
| regulation of macroautophagy | Any process that modulates the frequency, rate or extent of macroautophagy. |
| regulation of protein localization | Any process that modulates the frequency, rate or extent of any process in which a protein is transported to, or maintained in, a specific location. |
| response to mechanical stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| response to UV | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ultraviolet radiation (UV light) stimulus. Ultraviolet radiation is electromagnetic radiation with a wavelength in the range of 10 to 380 nanometers. |
| rhythmic process | Any process pertinent to the generation and maintenance of rhythms in the physiology of an organism. |
| stress-activated MAPK cascade | The series of molecular signals in which a stress-activated MAP kinase cascade relays a signal; MAP kinase cascades involve at least three protein kinase activities and culminate in the phosphorylation and activation of a MAP kinase. |
33 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32485 | HOG1 | Mitogen-activated protein kinase HOG1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P79996 | MAPK9 | Mitogen-activated protein kinase 9 | Gallus gallus (Chicken) | PR |
| P20794 | MAK | Serine/threonine-protein kinase MAK | Homo sapiens (Human) | PR |
| Q9UQ07 | MOK | MAPK/MAK/MRK overlapping kinase | Homo sapiens (Human) | PR |
| Q8TD08 | MAPK15 | Mitogen-activated protein kinase 15 | Homo sapiens (Human) | SS |
| P45984 | MAPK9 | Mitogen-activated protein kinase 9 | Homo sapiens (Human) | SS |
| P53779 | MAPK10 | Mitogen-activated protein kinase 10 | Homo sapiens (Human) | EV |
| Q9UBE8 | NLK | Serine/threonine-protein kinase NLK | Homo sapiens (Human) | SS |
| P27361 | MAPK3 | Mitogen-activated protein kinase 3 | Homo sapiens (Human) | SS |
| P28482 | MAPK1 | Mitogen-activated protein kinase 1 | Homo sapiens (Human) | EV |
| P31152 | MAPK4 | Mitogen-activated protein kinase 4 | Homo sapiens (Human) | SS |
| Q16659 | MAPK6 | Mitogen-activated protein kinase 6 | Homo sapiens (Human) | SS |
| Q13164 | MAPK7 | Mitogen-activated protein kinase 7 | Homo sapiens (Human) | SS |
| P53778 | MAPK12 | Mitogen-activated protein kinase 12 | Homo sapiens (Human) | SS |
| O15264 | MAPK13 | Mitogen-activated protein kinase 13 | Homo sapiens (Human) | SS |
| Q15759 | MAPK11 | Mitogen-activated protein kinase 11 | Homo sapiens (Human) | SS |
| Q16539 | MAPK14 | Mitogen-activated protein kinase 14 | Homo sapiens (Human) | SS |
| Q9WTU6 | Mapk9 | Mitogen-activated protein kinase 9 | Mus musculus (Mouse) | PR |
| Q91Y86 | Mapk8 | Mitogen-activated protein kinase 8 | Mus musculus (Mouse) | PR |
| P49186 | Mapk9 | Mitogen-activated protein kinase 9 | Rattus norvegicus (Rat) | PR |
| Q336X9 | MPK6 | Mitogen-activated protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q5J4W4 | MPK2 | Mitogen-activated protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84UI5 | MPK1 | Mitogen-activated protein kinase 1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10N20 | MPK5 | Mitogen-activated protein kinase 5 | Oryza sativa subsp. japonica (Rice) | SS |
| O44408 | kgb-1 | GLH-binding kinase 1 | Caenorhabditis elegans | PR |
| Q39023 | MPK3 | Mitogen-activated protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39026 | MPK6 | Mitogen-activated protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39025 | MPK5 | Mitogen-activated protein kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39024 | MPK4 | Mitogen-activated protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M1Z5 | MPK10 | Mitogen-activated protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GYQ5 | MPK12 | Mitogen-activated protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LMM5 | MPK11 | Mitogen-activated protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQQ9 | MPK13 | Mitogen-activated protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSRSKRDNNF | YSVEIGDSTF | TVLKRYQNLK | PIGSGAQGIV | CAAYDAILER | NVAIKKLSRP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FQNQTHAKRA | YRELVLMKCV | NHKNIIGLLN | VFTPQKSLEE | FQDVYIVMEL | MDANLCQVIQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MELDHERMSY | LLYQMLCGIK | HLHSAGIIHR | DLKPSNIVVK | SDCTLKILDF | GLARTAGTSF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MMTPYVVTRY | YRAPEVILGM | GYKENVDLWS | VGCIMGEMVC | HKILFPGRDY | IDQWNKVIEQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LGTPCPEFMK | KLQPTVRTYV | ENRPKYAGYS | FEKLFPDVLF | PADSEHNKLK | ASQARDLLSK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MLVIDASKRI | SVDEALQHPY | INVWYDPSEA | EAPPPKIPDK | QLDEREHTIE | EWKELIYKEV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MDLEERTKNG | VIRGQPSPLG | AAVINGSQHP | SSSSSVNDVS | SMSTDPTLAS | DTDSSLEAAA |
| GPLGCCR |