P43136
Gene name |
Nr2f6 (Ear-2, Ear2, Erbal2) |
Protein name |
Nuclear receptor subfamily 2 group F member 6 |
Names |
COUP transcription factor 3, COUP-TF3, V-erbA-related protein 2, EAR-2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:13864 |
EC number |
|
Protein Class |
|
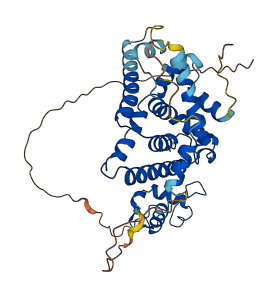
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P43136
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P43136-F1 | Predicted | AlphaFoldDB |
1 variants for P43136
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3399329690 | 87 | R>C | No | EVA |
No associated diseases with P43136
8 regional properties for P43136
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EF-hand domain | 188 - 216 | IPR002048-1 |
| domain | EF-hand domain | 304 - 339 | IPR002048-2 |
| domain | Mitochondrial Rho GTPase 1/3, EF hand associated, type-1 | 341 - 413 | IPR013566 |
| domain | EF hand associated, type-2 | 220 - 305 | IPR013567 |
| binding_site | EF-Hand 1, calcium-binding site | 197 - 209 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 317 - 329 | IPR018247-2 |
| domain | MIRO domain | 2 - 168 | IPR020860-1 |
| domain | MIRO domain | 416 - 579 | IPR020860-2 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| nuclear receptor activity | A DNA-binding transcription factor activity regulated by binding to a ligand that modulates the transcription of specific gene sets transcribed by RNA polymerase II. Nuclear receptor ligands are usually lipid-based (such as a steroid hormone) and the binding of the ligand to its receptor often occurs in the cytoplasm, which leads to its tranlocation to the nucleus. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| zinc ion binding | Binding to a zinc ion (Zn). |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure development | The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| detection of temperature stimulus involved in sensory perception of pain | The series of events involved in the perception of pain in which a temperature stimulus is received and converted into a molecular signal. |
| entrainment of circadian clock by photoperiod | The synchronization of a circadian rhythm to photoperiod, the intermittent cycle of light (day) and dark (night). |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| neuron development | The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9TTR7 | NR2F2 | COUP transcription factor 2 | Bos taurus (Bovine) | PR |
| O18971 | PPARG | Peroxisome proliferator-activated receptor gamma | Bos taurus (Bovine) | PR |
| Q90733 | NR2F2 | COUP transcription factor 2 | Gallus gallus (Chicken) | PR |
| P68306 | THRB | Thyroid hormone receptor beta | Gallus gallus (Chicken) | PR |
| A7X8B3 | PGR | Progesterone receptor | Pan troglodytes (Chimpanzee) | SS |
| P37231 | PPARG | Peroxisome proliferator-activated receptor gamma | Homo sapiens (Human) | PR |
| P06401 | PGR | Progesterone receptor | Homo sapiens (Human) | EV |
| P10589 | NR2F1 | COUP transcription factor 1 | Homo sapiens (Human) | PR |
| O75469 | NR1I2 | Nuclear receptor subfamily 1 group I member 2 | Homo sapiens (Human) | PR |
| P24468 | NR2F2 | COUP transcription factor 2 | Homo sapiens (Human) | PR |
| P49116 | NR2C2 | Nuclear receptor subfamily 2 group C member 2 | Homo sapiens (Human) | PR |
| P10588 | NR2F6 | Nuclear receptor subfamily 2 group F member 6 | Homo sapiens (Human) | PR |
| P35396 | Ppard | Peroxisome proliferator-activated receptor delta | Mus musculus (Mouse) | PR |
| Q00175 | Pgr | Progesterone receptor | Mus musculus (Mouse) | SS |
| Q9Z0Y9 | Nr1h3 | Oxysterols receptor LXR-alpha | Mus musculus (Mouse) | PR |
| P43135 | Nr2f2 | COUP transcription factor 2 | Mus musculus (Mouse) | PR |
| P49698 | Hnf4a | Hepatocyte nuclear factor 4-alpha | Mus musculus (Mouse) | PR |
| O62807 | PPARG | Peroxisome proliferator-activated receptor gamma | Sus scrofa (Pig) | PR |
| O09018 | Nr2f2 | COUP transcription factor 2 | Rattus norvegicus (Rat) | PR |
| Q63449 | Pgr | Progesterone receptor | Rattus norvegicus (Rat) | SS |
| Q8SQ01 | NR1I2 | Nuclear receptor subfamily 1 group I member 2 | Macaca mulatta (Rhesus macaque) | PR |
| G5EFF5 | daf-12 | Nuclear hormone receptor family member daf-12 | Caenorhabditis elegans | PR |
| O45460 | nhr-54 | Nuclear hormone receptor family member nhr-54 | Caenorhabditis elegans | PR |
| Q20765 | nhr-7 | Nuclear hormone receptor family member nhr-7 | Caenorhabditis elegans | PR |
| Q21006 | nhr-34 | Nuclear hormone receptor family member nhr-34 | Caenorhabditis elegans | PR |
| O17928 | nhr-52 | Nuclear hormone receptor family member nhr-52 | Caenorhabditis elegans | PR |
| Q21878 | nhr-1 | Nuclear hormone receptor family member nhr-1 | Caenorhabditis elegans | PR |
| O18141 | nhr-79 | Nuclear hormone receptor family member nhr-79 | Caenorhabditis elegans | PR |
| Q6PH18 | nr2f1b | Nuclear receptor subfamily 2 group F member 1-B | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAMVTGGWGD | PGGDTNGVDK | AGGSYPRATE | DDSASPPGAT | SDAEPGDEER | PGLQVDCVVC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GDKSSGKHYG | VFTCEGCKSF | FKRSIRRNLS | YTCRSNRDCQ | IDQHHRNQCQ | YCRLKKCFRV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GMRKEAVQRG | RIPHALPGPA | ACSPPGATGV | EPFTGPPVSE | LIAQLLRAEP | YPAAGRFGGG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GAVLGIDNVC | ELAARLLFST | VEWARHAPFF | PELPAADQVA | LLRLSWSELF | VLNAAQAALP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LHTAPLLAAA | GLHAAPMAAE | RAVAFMDQVR | AFQEQVDKLG | RLQVDAAEYG | CLKAIALFTP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DACGLSDPAH | VESLQEKAQV | ALTEYVRAQY | PSQPQRFGRL | LLRLPALRAV | PASLISQLFF |
| 370 | 380 | ||||
| MRLVGKTPIE | TLIRDMLLSG | STFNWPYGSG |