P42680
Gene name |
TEC (PSCTK4) |
Protein name |
Tyrosine-protein kinase Tec |
Names |
EC 2.7.10.2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:7006 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
|
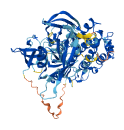
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
365-620 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
365-620 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
370-623 (Kinase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
506-530 (Activation loop from InterPro)
Target domain |
370-623 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Joseph RE et al. (2020) "Differential impact of BTK active site inhibitors on the conformational state of full-length BTK", eLife, 9,
- Kueffer LE et al. (2021) "Reining in BTK: Interdomain Interactions and Their Importance in the Regulatory Control of BTK", Frontiers in cell and developmental biology, 9, 655489
- Joseph RE et al. (2017) "Achieving a Graded Immune Response: BTK Adopts a Range of Active/Inactive Conformations Dictated by Multiple Interdomain Contacts", Structure (London, England : 1993), 25, 1481-1494.e4
- Wang Q et al. (2015) "Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate", eLife, 4,
- Nocka LM et al. (2023) "Stimulation of the catalytic activity of the tyrosine kinase Btk by the adaptor protein Grb2", eLife, 12,
- Lowe J et al. (2022) "Conformational switches that control the TEC kinase - PLCγ signaling axis", Journal of structural biology: X, 6, 100061
- Boyken SE et al. (2014) "A conserved isoleucine maintains the inactive state of Bruton's tyrosine kinase", Journal of molecular biology, 426, 3656-69
Autoinhibited structure

Activated structure

2 structures for P42680
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2LUL | NMR | - | A | 1-154 | PDB |
| AF-P42680-F1 | Predicted | AlphaFoldDB |
547 variants for P42680
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1723556805 | 5 | T>S | No | TOPMed | |
| rs1304801555 | 6 | I>V | No |
TOPMed gnomAD |
|
| rs1388197653 | 8 | E>K | No | gnomAD | |
| TCGA novel | 9 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1560418245 | 9 | E>V | No | Ensembl | |
| rs1560418242 | 10 | I>T | No | Ensembl | |
| rs1457600081 | 11 | L>P | No | gnomAD | |
| rs1395393224 | 14 | R>K | No | Ensembl | |
| rs1723555539 | 14 | R>W | No | TOPMed | |
| rs1304356128 | 15 | S>* | No |
TOPMed gnomAD |
|
| rs1345607054 | 16 | Q>P | No |
TOPMed gnomAD |
|
| rs759676495 | 17 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs1283283542 | 18 | K>E | No |
TOPMed gnomAD |
|
| rs776737918 | 18 | K>N | No |
ExAC gnomAD |
|
|
COSM1055801 rs766554666 |
19 | K>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs760902506 | 20 | K>N | No |
ExAC gnomAD |
|
| rs1723553613 | 20 | K>T | No | Ensembl | |
|
rs773480124 COSM1267597 |
21 | T>A | oesophagus [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
rs1723553284 COSM1267596 |
21 | T>I | oesophagus [Cosmic] | No |
cosmic curated TOPMed |
|
rs772497479 COSM3917872 |
22 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1322388850 | 25 | N>S | No | gnomAD | |
| rs769256758 | 26 | Y>C | No |
ExAC gnomAD |
|
| rs1723552288 | 27 | K>E | No | Ensembl | |
| rs748798649 | 27 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 28 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1055799 | 28 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779567477 | 29 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1435506923 | 29 | R>K | No |
TOPMed gnomAD |
|
| rs1390649123 | 30 | L>F | No |
TOPMed gnomAD |
|
| rs1291703382 | 31 | F>S | No | gnomAD | |
| rs1382362335 | 33 | L>F | No | gnomAD | |
| rs1457297072 | 34 | T>A | No | gnomAD | |
| rs148824532 | 34 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs148824532 | 34 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1328574336 | 36 | S>T | No | Ensembl | |
| rs745461310 | 37 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1166244762 | 37 | M>V | No | gnomAD | |
| rs145490815 | 39 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs759435324 | 40 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs759435324 | 40 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs143319990 | 41 | Y>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1723548915 | 42 | E>G | No |
TOPMed gnomAD |
|
| rs767837039 | 44 | R>* | No |
ExAC TOPMed gnomAD |
|
|
VAR_041850 rs35374286 |
44 | R>Q | No |
UniProt ExAC TOPMed dbSNP gnomAD |
|
| rs1723548067 | 45 | A>T | No | TOPMed | |
| rs1293641263 | 48 | K>N | No |
TOPMed gnomAD |
|
| rs1171685686 | 52 | G>A | No | gnomAD | |
| rs2109555637 | 52 | G>R | No | Ensembl | |
| rs1320770850 | 54 | I>S | No |
TOPMed gnomAD |
|
| rs1320770850 | 54 | I>T | No |
TOPMed gnomAD |
|
| TCGA novel | 55 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767902369 | 55 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs767902369 | 55 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1055939759 | 59 | I>V | No |
TOPMed gnomAD |
|
| rs1242089632 | 60 | K>Q | No |
TOPMed gnomAD |
|
| rs762078095 | 60 | K>R | No |
ExAC gnomAD |
|
| rs1253211646 | 61 | C>G | No | gnomAD | |
| rs1445679604 | 65 | V>M | No |
TOPMed gnomAD |
|
| rs1006089815 | 68 | D>G | No |
TOPMed gnomAD |
|
| rs1006089815 | 68 | D>V | No |
TOPMed gnomAD |
|
| rs1201307682 | 69 | D>G | No |
TOPMed gnomAD |
|
| rs1560391037 | 70 | G>A | No | gnomAD | |
| rs763388390 | 72 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1487535249 | 73 | P>A | No | TOPMed | |
| COSM6167515 | 73 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1721314034 | 74 | C>F | No | TOPMed | |
| rs776126020 | 74 | C>R | No |
ExAC gnomAD |
|
| TCGA novel | 76 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1288931917 | 79 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1721107856 | 82 | V>I | No | Ensembl | |
| rs1721107577 | 83 | V>I | No | TOPMed | |
| rs1721107461 | 84 | H>R | No | TOPMed | |
|
rs1180964661 COSM1637104 |
85 | D>E | bone [Cosmic] | No |
cosmic curated TOPMed |
| rs1721107182 | 85 | D>G | No | gnomAD | |
| rs1341844116 | 85 | D>N | No | gnomAD | |
| rs75574368 | 86 | A>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs75574368 | 86 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776146242 | 91 | I>V | No |
ExAC gnomAD |
|
| rs1721106096 | 93 | A>E | No | TOPMed | |
| COSM3974927 | 93 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1465973033 | 94 | P>A | No |
TOPMed gnomAD |
|
| rs1465973033 | 94 | P>S | No |
TOPMed gnomAD |
|
| COSM1429925 | 95 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs894500072 | 98 | S>N | No |
TOPMed gnomAD |
|
| TCGA novel | 100 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4901424 | 100 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1721105319 | 100 | D>N | No | TOPMed | |
| rs760332965 | 101 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1721105036 | 102 | W>* | No |
TOPMed gnomAD |
|
| rs1721104907 | 102 | W>* | No |
TOPMed gnomAD |
|
| rs940793291 | 104 | K>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 108 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1188210081 | 108 | E>G | No | gnomAD | |
| COSM4124960 | 109 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1721051550 | 109 | E>G | No | TOPMed | |
| TCGA novel | 110 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1483141979 | 110 | I>V | No | TOPMed | |
| COSM1055797 | 111 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772564593 | 112 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs767223534 | 112 | N>K | No |
ExAC gnomAD |
|
| rs774201318 | 117 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1342382369 | 117 | M>T | No | gnomAD | |
| TCGA novel | 118 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs911548228 | 122 | P>S | No | Ensembl | |
| rs200376782 | 126 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200376782 | 126 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1203145607 | 127 | D>E | No | gnomAD | |
| rs1275956051 | 129 | S>R | No | gnomAD | |
| rs1577718756 | 129 | S>T | No | Ensembl | |
| rs1407361042 | 131 | Q>K | No | gnomAD | |
| rs139186461 | 131 | Q>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs139186461 | 131 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1307457350 | 133 | C>Y | No | gnomAD | |
| COSM258159 | 137 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1410542142 | 138 | K>N | No |
TOPMed gnomAD |
|
| rs769826531 | 141 | P>A | No |
ExAC gnomAD |
|
| rs768480313 | 142 | G>R | No |
TOPMed gnomAD |
|
| COSM734279 | 143 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758673994 | 147 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1172955840 | 147 | N>S | No | gnomAD | |
| TCGA novel | 147 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1721047295 | 149 | F>L | No | TOPMed | |
| TCGA novel | 149 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1721047063 | 151 | S>I | No | TOPMed | |
| rs2271173 | 151 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1179621689 | 152 | S>N | No | gnomAD | |
| rs1720973261 | 153 | I>M | No |
TOPMed gnomAD |
|
| rs1271019174 | 153 | I>V | No |
TOPMed gnomAD |
|
| rs1720973136 | 154 | R>G | No | TOPMed | |
| rs1039903900 | 154 | R>K | No |
TOPMed gnomAD |
|
| rs370571771 | 156 | A>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs370571771 | 156 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1720972502 | 157 | L>V | No | TOPMed | |
| rs1474142023 | 158 | P>A | No |
TOPMed gnomAD |
|
| rs780358141 | 158 | P>L | No |
ExAC gnomAD |
|
| rs1474142023 | 158 | P>T | No |
TOPMed gnomAD |
|
| COSM481317 | 164 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756617928 | 165 | K>E | No |
ExAC gnomAD |
|
| rs1420726452 | 165 | K>N | No |
TOPMed gnomAD |
|
|
rs534338993 COSM1429923 |
166 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
| rs199797945 | 166 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs781592886 | 168 | P>L | No |
ExAC gnomAD |
|
|
rs1161019348 TCGA novel |
168 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1161019348 | 168 | P>T | No | TOPMed | |
| COSM734280 | 169 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 170 | P>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1389205450 | 170 | P>R | No | gnomAD | |
| rs1414796827 | 171 | P>S | No |
TOPMed gnomAD |
|
| rs1577716532 | 172 | I>T | No | Ensembl | |
| rs751140099 | 172 | I>V | No |
ExAC gnomAD |
|
| rs777374319 | 173 | P>L | No | ExAC | |
| rs1265850213 | 174 | L>I | No | gnomAD | |
| rs1488841792 | 175 | E>K | No | gnomAD | |
| rs1203118250 | 176 | E>G | No | gnomAD | |
| rs1271070833 | 176 | E>K | No | gnomAD | |
| COSM1429921 | 177 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs141394616 | 177 | E>D | No |
ESP ExAC gnomAD |
|
| rs752337530 | 179 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs764967387 | 180 | S>N | No |
ExAC gnomAD |
|
| rs1720937735 | 181 | E>G | No | TOPMed | |
| COSM1694356 | 181 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs574810323 | 183 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1285467499 | 184 | V>A | No | gnomAD | |
|
rs539573453 COSM1429920 |
184 | V>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2109542316 | 185 | V>I | No | Ensembl | |
| rs2109542304 | 186 | A>P | No | Ensembl | |
| rs1720936352 | 187 | M>I | No |
TOPMed gnomAD |
|
| rs1346070481 | 187 | M>T | No | gnomAD | |
| rs897275754 | 187 | M>V | No |
TOPMed gnomAD |
|
| rs760611263 | 189 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 189 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1720935830 | 191 | Q>H | No | Ensembl | |
| rs1720935969 | 191 | Q>L | No | Ensembl | |
| rs1407485278 | 192 | A>S | No |
TOPMed gnomAD |
|
| rs1407485278 | 192 | A>T | No |
TOPMed gnomAD |
|
| rs2109542251 | 193 | A>S | No | Ensembl | |
| rs1298434457 | 194 | E>A | No | gnomAD | |
| rs2109542243 | 194 | E>K | No | Ensembl | |
| rs768664754 | 195 | G>E | No |
ExAC TOPMed gnomAD |
|
|
rs768664754 COSM359910 |
195 | G>V | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1376345712 | 196 | H>R | No |
TOPMed gnomAD |
|
| rs1239721322 | 198 | L>V | No |
TOPMed gnomAD |
|
| rs763008568 | 199 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1432122009 | 200 | L>* | No | gnomAD | |
| COSM1310099 | 201 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370906002 | 202 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs746270033 | 202 | R>S | No |
ExAC gnomAD |
|
| rs1720932409 | 203 | G>D | No |
TOPMed gnomAD |
|
| rs941365394 | 204 | Q>E | No |
TOPMed gnomAD |
|
| rs771376138 | 206 | Y>* | No |
ExAC gnomAD |
|
| rs1482405725 | 206 | Y>C | No | gnomAD | |
| rs1197835955 | 206 | Y>D | No |
TOPMed gnomAD |
|
| TCGA novel | 206 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1720930800 | 207 | L>F | No | Ensembl | |
| rs1003501745 | 208 | I>T | No |
TOPMed gnomAD |
|
| COSM1055795 | 210 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777431361 | 210 | E>K | No |
ExAC gnomAD |
|
|
COSM1055793 rs905849900 |
211 | K>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs1226010019 | 213 | D>E | No | gnomAD | |
| TCGA novel | 213 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2109542108 | 214 | V>A | No | Ensembl | |
| rs2109542115 | 214 | V>I | No | Ensembl | |
| rs1335722528 | 215 | H>P | No | gnomAD | |
| rs1309211010 | 215 | H>Q | No |
TOPMed gnomAD |
|
| COSM6167516 | 216 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs566875728 | 216 | W>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747656157 | 217 | W>C | No |
ExAC gnomAD |
|
| rs1023856881 | 218 | R>G | No |
TOPMed gnomAD |
|
| rs1399616619 | 218 | R>K | No |
TOPMed gnomAD |
|
| rs1399616619 | 218 | R>T | No |
TOPMed gnomAD |
|
| rs1560385830 | 221 | D>G | No | Ensembl | |
| rs1302593702 | 221 | D>N | No |
TOPMed gnomAD |
|
| rs2109542062 | 224 | G>V | No | Ensembl | |
| rs779559749 | 226 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1341640879 | 227 | G>V | No | Ensembl | |
| rs1272263155 | 231 | S>N | No |
TOPMed gnomAD |
|
|
COSM288307 rs750190240 |
234 | V>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs191626703 | 235 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1289506269 | 237 | K>N | No |
TOPMed gnomAD |
|
| COSM1055791 | 238 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757140518 | 239 | S>P | No |
ExAC gnomAD |
|
| rs752755232 | 240 | N>D | No |
ExAC gnomAD |
|
| rs973959312 | 240 | N>S | No |
TOPMed gnomAD |
|
| rs1242626865 | 241 | N>H | No | gnomAD | |
| rs1195314878 | 243 | D>N | No |
TOPMed gnomAD |
|
| rs1195314878 | 243 | D>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 245 | Y>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1387746424 | 246 | E>K | No | TOPMed | |
| rs1476975842 | 247 | W>* | No | gnomAD | |
| rs1428541566 | 249 | C>F | No | gnomAD | |
| rs1039698473 | 252 | M>I | No | TOPMed | |
| rs1720418920 | 252 | M>V | No | TOPMed | |
| rs1490707890 | 253 | N>K | No | gnomAD | |
| rs779798762 | 253 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs138464079 | 255 | S>N | No |
ESP ExAC gnomAD |
|
| rs745689320 | 256 | K>R | No |
ExAC gnomAD |
|
| rs1438063005 | 257 | A>E | No | gnomAD | |
| COSM6100376 | 258 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1323393088 | 260 | L>V | No | gnomAD | |
| rs147079314 | 262 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs137884397 | 262 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs137884397 | 262 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200509274 | 263 | S>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 264 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3825928 rs761984494 |
269 | G>D | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs764686962 | 271 | M>V | No |
ExAC gnomAD |
|
| rs1035732920 | 272 | V>I | No | Ensembl | |
| COSM587369 | 274 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1409664773 | 275 | S>F | No |
TOPMed gnomAD |
|
| rs1409664773 | 275 | S>Y | No |
TOPMed gnomAD |
|
| rs2109516794 | 276 | S>C | No | Ensembl | |
| COSM3825927 | 277 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1443645002 | 278 | P>S | No |
TOPMed gnomAD |
|
| rs1443645002 | 278 | P>T | No |
TOPMed gnomAD |
|
| rs750310314 | 282 | T>I | No |
ExAC gnomAD |
|
| rs1213280873 | 282 | T>P | No | gnomAD | |
| rs867297832 | 284 | S>Y | No | Ensembl | |
| rs2109516741 | 285 | L>F | No | Ensembl | |
| rs1720130529 | 287 | T>A | No | TOPMed | |
| rs1463895029 | 287 | T>I | No | gnomAD | |
| rs1260702660 | 288 | K>R | No | gnomAD | |
| rs759030525 | 290 | G>E | No |
ExAC gnomAD |
|
| rs764277354 | 291 | G>E | No | gnomAD | |
| rs1322673151 | 292 | E>D | No |
TOPMed gnomAD |
|
| rs1332172366 | 292 | E>G | No | TOPMed | |
| rs776093286 | 293 | G>A | No |
ExAC gnomAD |
|
| rs776093286 | 293 | G>D | No |
ExAC gnomAD |
|
| rs776093286 | 293 | G>V | No |
ExAC gnomAD |
|
| COSM3428540 | 294 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs61729380 | 294 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs773063384 | 295 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs2109515036 | 296 | G>D | No | Ensembl | |
| rs747952759 | 296 | G>S | No |
ExAC gnomAD |
|
| rs527854494 | 298 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1720074778 | 300 | Y>* | No | gnomAD | |
| rs1316209550 | 300 | Y>C | No | TOPMed | |
| rs768694592 | 301 | H>Q | No | ExAC | |
| rs541078451 | 306 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs541078451 | 306 | T>R | No |
1000Genomes ExAC gnomAD |
|
| rs757509359 | 307 | T>A | No |
ExAC gnomAD |
|
| rs757509359 | 307 | T>S | No |
ExAC gnomAD |
|
| rs1490545987 | 308 | S>C | No | gnomAD | |
| rs1490545987 | 308 | S>Y | No | gnomAD | |
| rs561711226 | 311 | K>N | No |
1000Genomes ExAC gnomAD |
|
| rs543420319 | 318 | H>P | No |
1000Genomes ExAC gnomAD |
|
| rs543420319 | 318 | H>R | No |
1000Genomes ExAC gnomAD |
|
| rs1560376063 | 322 | S>P | No | Ensembl | |
| rs1720071958 | 323 | I>V | No | TOPMed | |
|
rs1309161559 COSM1310098 |
325 | E>D | urinary_tract [Cosmic] | No |
cosmic curated gnomAD |
| rs765779795 | 325 | E>K | No |
ExAC gnomAD |
|
| rs576422071 | 326 | I>V | No |
1000Genomes ExAC gnomAD |
|
| rs1345656048 | 327 | I>T | No |
TOPMed gnomAD |
|
| COSM285650 | 327 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM6100377 rs746395396 COSM587370 |
328 | E>D | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1720069933 | 329 | Y>H | No | Ensembl | |
| rs1169580855 | 331 | K>N | No | TOPMed | |
| rs1720069315 | 334 | A>V | No | Ensembl | |
| rs1344147450 | 336 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1408957123 | 340 | R>G | No | TOPMed | |
| rs747111814 | 341 | L>V | No |
ExAC gnomAD |
|
| rs777892454 | 342 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1453194718 | 342 | R>W | No | gnomAD | |
| rs748532219 | 343 | Y>N | No |
ExAC TOPMed gnomAD |
|
| rs779206975 | 344 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1240965997 | 344 | P>R | No | gnomAD | |
| rs754284170 | 345 | V>I | No |
ExAC gnomAD |
|
| rs780597024 | 346 | S>R | No |
ExAC gnomAD |
|
| rs1265035864 | 347 | V>A | No |
TOPMed gnomAD |
|
| rs1265035864 | 347 | V>E | No |
TOPMed gnomAD |
|
| rs1202827426 | 351 | N>D | No | gnomAD | |
| COSM3917870 | 351 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs151151021 | 351 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1560374359 | 352 | A>T | No | Ensembl | |
| rs761515779 | 353 | P>L | No |
ExAC gnomAD |
|
| rs767097402 | 353 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs763921020 | 357 | G>E | No |
ExAC gnomAD |
|
| TCGA novel | 359 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1168086018 COSM1055789 |
361 | E>D | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 361 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1719873859 | 363 | W>* | No |
TOPMed gnomAD |
|
| rs1719873984 | 363 | W>R | No | gnomAD | |
| rs1719873721 | 364 | E>K | No | Ensembl | |
| rs144659420 | 367 | P>A | No |
ESP ExAC |
|
| TCGA novel | 367 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765113539 | 370 | L>R | No | ExAC | |
| rs565655073 | 370 | L>V | No | Ensembl | |
| rs946733895 | 374 | R>S | No | gnomAD | |
| rs201608922 | 376 | L>S | No |
ExAC gnomAD |
|
| rs868276572 | 377 | G>R | No | Ensembl | |
| rs140868704 | 378 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs767529253 | 378 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs761869739 | 380 | L>P | No |
ExAC gnomAD |
|
| TCGA novel | 381 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1271268431 | 383 | V>A | No | gnomAD | |
| rs1719870743 | 383 | V>M | No | gnomAD | |
| rs749636109 | 385 | R>K | No |
ExAC gnomAD |
|
| COSM481315 | 385 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1280697465 | 386 | L>P | No | gnomAD | |
| rs199646567 | 389 | W>R | No |
ExAC TOPMed gnomAD |
|
| rs1254880258 | 390 | R>* | No | gnomAD | |
| rs746326176 | 390 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1719868798 | 391 | A>T | No | gnomAD | |
| rs1312156368 | 392 | Q>K | No | TOPMed | |
| rs756890387 | 395 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1353010758 | 395 | V>L | No | gnomAD | |
| rs752469056 | 396 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1228858 rs752469056 |
396 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| COSM3825926 | 397 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765018274 | 398 | K>R | No |
ExAC TOPMed gnomAD |
|
|
rs1484539226 COSM587371 COSM6100378 |
401 | R>L | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1484539226 | 401 | R>Q | No | gnomAD | |
| rs754887835 | 401 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1719865994 | 402 | E>G | No | Ensembl | |
| COSM734281 | 403 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1199548264 | 404 | A>S | No | gnomAD | |
| rs1719865272 | 405 | M>I | No | TOPMed | |
| rs1719865414 | 405 | M>T | No | Ensembl | |
| rs1230873045 | 405 | M>V | No | gnomAD | |
| rs961818658 | 406 | C>* | No |
TOPMed gnomAD |
|
| rs961818658 | 406 | C>W | No |
TOPMed gnomAD |
|
|
COSM1429918 rs1719864805 |
407 | E>K | large_intestine [Cosmic] | No |
cosmic curated TOPMed |
| rs1438827121 | 408 | E>* | No | gnomAD | |
| rs868616029 | 411 | I>K | No | Ensembl | |
| rs868616029 | 411 | I>T | No | Ensembl | |
| rs1260541310 | 414 | A>T | No | gnomAD | |
| rs774365822 | 415 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs764331227 | 416 | V>M | No |
ExAC gnomAD |
|
| rs41307144 | 418 | M>R | No | TOPMed | |
| rs41307144 | 418 | M>T | No | TOPMed | |
| rs762925756 | 419 | K>T | No |
ExAC gnomAD |
|
| rs369398035 | 423 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs369398035 | 423 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs145617527 | 426 | V>M | No |
ESP gnomAD |
|
| rs1719855761 | 428 | L>F | No | Ensembl | |
| rs999385114 | 429 | Y>S | No | Ensembl | |
| rs777091415 | 431 | V>A | No |
ExAC gnomAD |
|
| rs777091415 | 431 | V>E | No |
ExAC gnomAD |
|
| rs1186478879 | 433 | T>S | No |
TOPMed gnomAD |
|
| rs555121105 | 433 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs773014819 | 434 | Q>* | No | Ensembl | |
| COSM3604277 | 437 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772865397 | 437 | P>R | No |
ExAC gnomAD |
|
| rs1241549571 | 438 | I>T | No | gnomAD | |
| rs61744269 | 442 | T>I | No | Ensembl | |
| rs1164902930 | 445 | M>V | No | gnomAD | |
| rs149757303 | 446 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1719853568 | 446 | E>K | No | TOPMed | |
| rs1163619725 | 447 | R>K | No | Ensembl | |
| rs1560373439 | 448 | G>V | No | Ensembl | |
| rs559961259 | 450 | L>F | No | 1000Genomes | |
| rs1719852671 | 453 | F>S | No | gnomAD | |
| COSM3604275 | 454 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs768171223 COSM247775 |
455 | R>* | prostate [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs372216578 | 455 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs75184130 | 457 | R>G | No | Ensembl | |
| rs374511784 | 458 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1719851613 | 459 | G>D | No | Ensembl | |
| rs750417190 | 460 | H>D | No |
ExAC gnomAD |
|
| rs1719851361 | 462 | S>R | No | Ensembl | |
| rs780984861 | 465 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs780984861 | 465 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs752693893 | 468 | S>G | No |
ExAC gnomAD |
|
| COSM4124958 | 468 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1560373331 | 469 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs765514796 | 470 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1680429350 | 473 | V>M | No | TOPMed | |
| rs1427207673 | 474 | C>Y | No | gnomAD | |
| rs759796647 | 475 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1194437456 | 476 | G>E | No |
TOPMed gnomAD |
|
| rs1194437456 | 476 | G>V | No |
TOPMed gnomAD |
|
| rs1719849351 | 477 | M>I | No | gnomAD | |
| COSM6167517 | 478 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754263100 | 478 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs761110899 | 479 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs773925325 | 480 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs139680290 | 481 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs910063103 | 483 | N>D | No | Ensembl | |
| rs761436897 | 483 | N>T | No |
ExAC gnomAD |
|
| COSM6167518 | 484 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1223527921 | 484 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs773686592 | 484 | S>R | No |
ExAC gnomAD |
|
| rs1332465750 | 497 | V>A | No |
TOPMed gnomAD |
|
| rs1719658017 | 498 | S>N | No | gnomAD | |
|
rs763650133 COSM1429917 |
500 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1719657050 | 501 | G>R | No | TOPMed | |
| rs775160457 | 502 | V>I | No |
ExAC gnomAD |
|
| rs769412539 | 505 | V>I | No |
ExAC TOPMed gnomAD |
|
|
rs150324512 COSM109828 |
509 | G>E | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs1256817357 | 509 | G>R | No | TOPMed | |
| COSM4895307 | 510 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1237293687 | 510 | M>K | No |
TOPMed gnomAD |
|
| rs1237293687 | 510 | M>T | No |
TOPMed gnomAD |
|
| rs145358315 | 514 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1012041959 | 517 | D>N | No |
TOPMed gnomAD |
|
| rs1012041959 | 517 | D>Y | No |
TOPMed gnomAD |
|
| rs896582721 | 520 | T>I | No | Ensembl | |
| rs1283975092 | 520 | T>P | No | gnomAD | |
| rs1246113513 | 521 | S>G | No | gnomAD | |
| COSM447908 | 522 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs369215066 | 524 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1381762842 | 526 | K>E | No |
TOPMed gnomAD |
|
| rs1381762842 | 526 | K>Q | No |
TOPMed gnomAD |
|
| rs1422989284 | 527 | F>L | No | gnomAD | |
| rs1300343530 | 527 | F>L | No |
TOPMed gnomAD |
|
| rs1363412879 | 529 | V>G | No | gnomAD | |
| COSM3917869 | 530 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746818910 | 532 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs139947477 | 534 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1479230692 | 536 | V>M | No | Ensembl | |
| rs200292050 | 539 | Y>F | No |
TOPMed gnomAD |
|
| rs200292050 | 539 | Y>S | No |
TOPMed gnomAD |
|
| TCGA novel | 540 | S>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376411203 | 541 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs746133746 | 541 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs746133746 | 541 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs746133746 | 541 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs751891388 | 544 | S>G | No | Ensembl | |
|
COSM2149549 rs1210858990 |
544 | S>N | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs757601498 | 546 | S>* | No |
ExAC gnomAD |
|
| COSM1310097 | 546 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1719545755 | 547 | D>E | No | gnomAD | |
| rs1719545405 | 548 | V>G | No | Ensembl | |
| rs1250190269 | 548 | V>I | No | gnomAD | |
| rs113970135 | 549 | W>R | No | Ensembl | |
| rs147484158 | 552 | G>A | No |
ESP TOPMed |
|
| rs147484158 | 552 | G>D | No |
ESP TOPMed |
|
| rs1363722534 | 553 | V>A | No | Ensembl | |
| rs1719540036 | 553 | V>F | No | gnomAD | |
| rs1268288282 | 554 | L>S | No | gnomAD | |
| rs769831718 | 555 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs532438469 | 555 | M>L | No | gnomAD | |
| rs1211914902 | 555 | M>T | No | gnomAD | |
| rs532438469 | 555 | M>V | No | gnomAD | |
| rs781315048 | 556 | W>G | No |
ExAC gnomAD |
|
| rs781315048 | 556 | W>R | No |
ExAC gnomAD |
|
| rs1719538882 | 558 | V>L | No | Ensembl | |
|
COSM1228854 rs185446950 |
560 | T>M | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
COSM1540311 COSM6167519 rs1719538623 |
560 | T>S | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic TOPMed |
| rs1023904247 | 561 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 562 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_041851 | 563 | R>K | a lung adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
| COSM3775874 | 563 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1014141563 | 564 | M>I | No | TOPMed | |
| rs940417359 | 565 | P>R | No | Ensembl | |
| rs1719537610 | 566 | F>S | No | TOPMed | |
| rs1719536985 | 570 | T>N | No | Ensembl | |
| rs1401001159 | 571 | N>S | No |
TOPMed gnomAD |
|
| rs758922575 | 574 | V>A | No |
ExAC gnomAD |
|
| rs907574224 | 575 | V>I | No |
TOPMed gnomAD |
|
| rs1719536157 | 576 | T>S | No | TOPMed | |
| rs1424013141 | 577 | M>T | No | gnomAD | |
| rs1719536030 | 577 | M>V | No | Ensembl | |
| rs1719535762 | 578 | V>A | No | TOPMed | |
| rs1478808162 | 579 | T>I | No |
TOPMed gnomAD |
|
| rs779390935 | 580 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs182366408 | 580 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs182366408 | 580 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs753368684 | 582 | H>Y | No |
ExAC gnomAD |
|
|
rs765971294 COSM1055787 |
583 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs760335372 | 584 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1719533187 | 585 | Y>C | No | TOPMed | |
| rs952056934 | 586 | Q>R | No |
TOPMed gnomAD |
|
| rs761626081 | 587 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs761626081 | 587 | P>Q | No |
ExAC gnomAD |
|
|
COSM6167520 COSM1540312 rs1306776603 |
588 | K>N | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs148361115 | 589 | L>V | No |
ESP TOPMed gnomAD |
|
| rs1719531964 | 589 | L>W | No | Ensembl | |
| rs1377544625 | 590 | A>T | No | gnomAD | |
| rs761695684 | 590 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs149649815 | 591 | S>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs759553648 | 592 | N>D | No |
ExAC gnomAD |
|
| TCGA novel | 592 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1415775291 | 592 | N>S | No | gnomAD | |
| rs776729923 | 593 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs771059101 | 594 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1158386165 | 594 | V>M | No | gnomAD | |
| rs960602007 | 595 | Y>C | No | Ensembl | |
| rs747172604 | 596 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1719529551 | 598 | M>I | No | TOPMed | |
| rs1719529413 | 599 | L>P | No | Ensembl | |
| rs1212392632 | 602 | W>* | No | gnomAD | |
| rs1445462271 | 604 | E>D | No |
TOPMed gnomAD |
|
| rs1719475672 | 606 | P>L | No | TOPMed | |
| rs766542747 | 606 | P>S | No |
ExAC gnomAD |
|
| rs760741758 | 607 | E>G | No |
ExAC TOPMed |
|
| rs748536244 | 610 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs748536244 | 610 | P>T | No |
ExAC TOPMed gnomAD |
|
|
COSM3365652 rs1560368654 |
611 | S>Y | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs552094470 | 612 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1464522425 | 613 | E>G | No | gnomAD | |
| rs1027163742 | 613 | E>K | No |
TOPMed gnomAD |
|
| rs138487528 | 617 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
rs533445085 COSM1228856 |
617 | R>H | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs375268361 | 618 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA |
| rs1269931727 | 618 | T>I | No | gnomAD | |
| rs1247084546 | 619 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1488069401 | 619 | I>V | No | gnomAD | |
| rs781013041 | 620 | D>H | No |
ExAC gnomAD |
|
| TCGA novel | 621 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1324007287 | 623 | V>I | No |
TOPMed gnomAD |
|
| rs1324007287 | 623 | V>L | No |
TOPMed gnomAD |
|
| rs1316027340 | 625 | C>S | No | gnomAD | |
| rs756926914 | 625 | C>Y | No |
ExAC gnomAD |
|
| rs763889694 | 627 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs373626137 | 627 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 629 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1003226298 | 632 | R>Q | No | Ensembl |
No associated diseases with P42680
10 regional properties for P42680
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 370 - 623 | IPR000719 |
| domain | SH2 domain | 245 - 345 | IPR000980 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 370 - 618 | IPR001245 |
| domain | SH3 domain | 179 - 239 | IPR001452 |
| conserved_site | Zinc finger, Btk motif | 102 - 149 | IPR001562 |
| domain | Pleckstrin homology domain | 4 - 113 | IPR001849 |
| active_site | Tyrosine-protein kinase, active site | 485 - 497 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 376 - 398 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 370 - 619 | IPR020635 |
| domain | Tyrosine-protein kinase Tec, SH3 domain | 182 - 237 | IPR035572 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction |
| phospholipid binding | Binding to a phospholipid, a class of lipids containing phosphoric acid as a mono- or diester. |
| protein self-association | Binding to a domain within the same polypeptide. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigens produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| B cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a B cell. |
| integrin-mediated signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to an integrin on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of platelet activation | Any process that modulates the rate or frequency of platelet activation. Platelet activation is a series of progressive, overlapping events triggered by exposure of the platelets to subendothelial tissue. |
| T cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a T cell. |
| tissue regeneration | The regrowth of lost or destroyed tissues. |
86 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0JNB0 | FYN | Tyrosine-protein kinase Fyn | Bos taurus (Bovine) | SS |
| Q0VBZ0 | CSK | Tyrosine-protein kinase CSK | Bos taurus (Bovine) | SS |
| Q3ZC95 | BTK | Tyrosine-protein kinase | Bos taurus (Bovine) | EV SS |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| P00523 | SRC | Proto-oncogene tyrosine-protein kinase Src | Gallus gallus (Chicken) | EV |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| Q75R65 | JAK2 | Tyrosine-protein kinase JAK2 | Gallus gallus (Chicken) | SS |
| Q24592 | hop | Tyrosine-protein kinase hopscotch | Drosophila melanogaster (Fruit fly) | PR |
| Q9V9J3 | Src42A | Tyrosine-protein kinase Src42A | Drosophila melanogaster (Fruit fly) | SS |
| P00528 | Src64B | Tyrosine-protein kinase Src64B | Drosophila melanogaster (Fruit fly) | SS |
| P08630 | Btk | Tyrosine-protein kinase Btk | Drosophila melanogaster (Fruit fly) | SS |
| P23458 | JAK1 | Tyrosine-protein kinase JAK1 | Homo sapiens (Human) | SS |
| O60674 | JAK2 | Tyrosine-protein kinase JAK2 | Homo sapiens (Human) | EV |
| P52333 | JAK3 | Tyrosine-protein kinase JAK3 | Homo sapiens (Human) | SS |
| P29597 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | Homo sapiens (Human) | EV |
| P07947 | YES1 | Tyrosine-protein kinase Yes | Homo sapiens (Human) | SS |
| P09769 | FGR | Tyrosine-protein kinase Fgr | Homo sapiens (Human) | SS |
| P41240 | CSK | Tyrosine-protein kinase CSK | Homo sapiens (Human) | SS |
| P51451 | BLK | Tyrosine-protein kinase Blk | Homo sapiens (Human) | SS |
| P51813 | BMX | Cytoplasmic tyrosine-protein kinase BMX | Homo sapiens (Human) | SS |
| Q08881 | ITK | Tyrosine-protein kinase ITK/TSK | Homo sapiens (Human) | EV |
| P43405 | SYK | Tyrosine-protein kinase SYK | Homo sapiens (Human) | EV |
| P43403 | ZAP70 | Tyrosine-protein kinase ZAP-70 | Homo sapiens (Human) | EV |
| Q13882 | PTK6 | Protein-tyrosine kinase 6 | Homo sapiens (Human) | EV |
| P07948 | LYN | Tyrosine-protein kinase Lyn | Homo sapiens (Human) | SS |
| P06241 | FYN | Tyrosine-protein kinase Fyn | Homo sapiens (Human) | SS |
| P12931 | SRC | Proto-oncogene tyrosine-protein kinase Src | Homo sapiens (Human) | EV |
| P06239 | LCK | Tyrosine-protein kinase Lck | Homo sapiens (Human) | EV |
| P08631 | HCK | Tyrosine-protein kinase HCK | Homo sapiens (Human) | EV |
| P42685 | FRK | Tyrosine-protein kinase FRK | Homo sapiens (Human) | EV |
| Q06187 | BTK | Tyrosine-protein kinase BTK | Homo sapiens (Human) | EV |
| P42679 | MATK | Megakaryocyte-associated tyrosine-protein kinase | Homo sapiens (Human) | SS |
| Q14289 | PTK2B | Protein-tyrosine kinase 2-beta | Homo sapiens (Human) | PR |
| Q05397 | PTK2 | Focal adhesion kinase 1 | Homo sapiens (Human) | EV |
| Q13470 | TNK1 | Non-receptor tyrosine-protein kinase TNK1 | Homo sapiens (Human) | PR |
| Q07912 | TNK2 | Activated CDC42 kinase 1 | Homo sapiens (Human) | EV |
| P16591 | FER | Tyrosine-protein kinase Fer | Homo sapiens (Human) | PR |
| Q6J9G0 | STYK1 | Tyrosine-protein kinase STYK1 | Homo sapiens (Human) | PR |
| P42684 | ABL2 | Tyrosine-protein kinase ABL2 | Homo sapiens (Human) | SS |
| P00519 | ABL1 | Tyrosine-protein kinase ABL1 | Homo sapiens (Human) | EV |
| Q9R117 | Tyk2 | Non-receptor tyrosine-protein kinase TYK2 | Mus musculus (Mouse) | SS |
| P08103 | Hck | Tyrosine-protein kinase HCK | Mus musculus (Mouse) | SS |
| P16277 | Blk | Tyrosine-protein kinase Blk | Mus musculus (Mouse) | SS |
| Q62270 | Srms | Tyrosine-protein kinase Srms | Mus musculus (Mouse) | SS |
| Q64434 | Ptk6 | Protein-tyrosine kinase 6 | Mus musculus (Mouse) | SS |
| P05480 | Src | Proto-oncogene tyrosine-protein kinase Src | Mus musculus (Mouse) | EV |
| P14234 | Fgr | Tyrosine-protein kinase Fgr | Mus musculus (Mouse) | SS |
| P35991 | Btk | Tyrosine-protein kinase BTK | Mus musculus (Mouse) | EV |
| P41241 | Csk | Tyrosine-protein kinase CSK | Mus musculus (Mouse) | EV |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | Mus musculus (Mouse) | EV |
| Q62137 | Jak3 | Tyrosine-protein kinase JAK3 | Mus musculus (Mouse) | SS |
| Q62120 | Jak2 | Tyrosine-protein kinase JAK2 | Mus musculus (Mouse) | EV |
| P06240 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Mus musculus (Mouse) | SS |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | Mus musculus (Mouse) | SS |
| P39688 | Fyn | Tyrosine-protein kinase Fyn | Mus musculus (Mouse) | SS |
| P52332 | Jak1 | Tyrosine-protein kinase JAK1 | Mus musculus (Mouse) | SS |
| Q03526 | Itk | Tyrosine-protein kinase ITK/TSK | Mus musculus (Mouse) | SS |
| P41242 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q922K9 | Frk | Tyrosine-protein kinase FRK | Mus musculus (Mouse) | SS |
| P24604 | Tec | Tyrosine-protein kinase Tec | Mus musculus (Mouse) | SS |
| A1Y2K1 | FYN | Tyrosine-protein kinase Fyn | Sus scrofa (Pig) | SS |
| O19064 | JAK2 | Tyrosine-protein kinase JAK2 | Sus scrofa (Pig) | SS |
| Q62662 | Frk | Tyrosine-protein kinase FRK | Rattus norvegicus (Rat) | SS |
| Q62844 | Fyn | Tyrosine-protein kinase Fyn | Rattus norvegicus (Rat) | SS |
| Q07014 | Lyn | Tyrosine-protein kinase Lyn | Rattus norvegicus (Rat) | SS |
| P50545 | Hck | Tyrosine-protein kinase HCK | Rattus norvegicus (Rat) | SS |
| Q9WUD9 | Src | Proto-oncogene tyrosine-protein kinase Src | Rattus norvegicus (Rat) | SS |
| Q01621 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Rattus norvegicus (Rat) | SS |
| Q6P6U0 | Fgr | Tyrosine-protein kinase Fgr | Rattus norvegicus (Rat) | SS |
| Q62689 | Jak2 | Tyrosine-protein kinase JAK2 | Rattus norvegicus (Rat) | SS |
| Q63272 | Jak3 | Tyrosine-protein kinase JAK3 | Rattus norvegicus (Rat) | SS |
| P32577 | Csk | Tyrosine-protein kinase CSK | Rattus norvegicus (Rat) | SS |
| P41243 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Rattus norvegicus (Rat) | SS |
| F1LM93 | Yes1 | Tyrosine-protein kinase Yes | Rattus norvegicus (Rat) | SS |
| O45539 | src-2 | Tyrosine protein-kinase src-2 | Caenorhabditis elegans | SS |
| G5ECJ6 | csk-1 | Tyrosine-protein kinase csk-1 | Caenorhabditis elegans | SS |
| G5EE56 | src-1 | Tyrosine protein-kinase src-1 | Caenorhabditis elegans | SS |
| A1A5H8 | yes1 | Tyrosine-protein kinase yes | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F1RDG9 | fynb | Tyrosine-protein kinase fynb | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O12990 | jak1 | Tyrosine-protein kinase JAK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q1JPZ3 | src | Proto-oncogene tyrosine-protein kinase Src | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6EWH2 | fyna | Tyrosine-protein kinase fyna | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNFNTILEEI | LIKRSQQKKK | TSPLNYKERL | FVLTKSMLTY | YEGRAEKKYR | KGFIDVSKIK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CVEIVKNDDG | VIPCQNKYPF | QVVHDANTLY | IFAPSPQSRD | LWVKKLKEEI | KNNNNIMIKY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| HPKFWTDGSY | QCCRQTEKLA | PGCEKYNLFE | SSIRKALPPA | PETKKRRPPP | PIPLEEEDNS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EEIVVAMYDF | QAAEGHDLRL | ERGQEYLILE | KNDVHWWRAR | DKYGNEGYIP | SNYVTGKKSN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NLDQYEWYCR | NMNRSKAEQL | LRSEDKEGGF | MVRDSSQPGL | YTVSLYTKFG | GEGSSGFRHY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HIKETTTSPK | KYYLAEKHAF | GSIPEIIEYH | KHNAAGLVTR | LRYPVSVKGK | NAPTTAGFSY |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EKWEINPSEL | TFMRELGSGL | FGVVRLGKWR | AQYKVAIKAI | REGAMCEEDF | IEEAKVMMKL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| THPKLVQLYG | VCTQQKPIYI | VTEFMERGCL | LNFLRQRQGH | FSRDVLLSMC | QDVCEGMEYL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ERNSFIHRDL | AARNCLVSEA | GVVKVSDFGM | ARYVLDDQYT | SSSGAKFPVK | WCPPEVFNYS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RFSSKSDVWS | FGVLMWEVFT | EGRMPFEKYT | NYEVVTMVTR | GHRLYQPKLA | SNYVYEVMLR |
| 610 | 620 | 630 | |||
| CWQEKPEGRP | SFEDLLRTID | ELVECEETFG | R |