P41743
Gene name |
PRKCI (DXS1179E) |
Protein name |
Protein kinase C iota type |
Names |
Atypical protein kinase C-lambda/iota, PRKC-lambda/iota, aPKC-lambda/iota, nPKC-iota |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5584 |
EC number |
|
Protein Class |
RIBOSOMAL PROTEIN S6 KINASE (PTHR24351) |
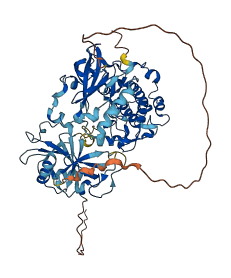
Descriptions
PRKCI, an atypical PKC (aPKC) isoform, is an important kinase in multiple cellular processes and regulated by the phosphatidylinositol 3-kinase product phosphatidylinositol 3,4,5-(PO4)3 (PIP3). In aPKC molecule, the arginine motif (123-140) in the pseudosubstrate region on the regulatory domain binds to acidic residues in the substrate-binding region on the catalytic domain, leading to the off state of aPKCs. The autoinhibition of aPKC is released by binding with the acidic ligand PIP3 and consequently exposing the substrate-binding site.
Autoinhibitory domains (AIDs)
Target domain |
233-596 (Catalytic domain of the Serine/Threonine Kinase, Atypical Protein Kinase C iota) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment, Deletion assay, Structural analysis |
Accessory elements
395-418 (Activation loop from InterPro)
Target domain |
233-596 (Catalytic domain of the Serine/Threonine Kinase, Atypical Protein Kinase C iota) |
Relief mechanism |
|
Assay |
|
References
- Ivey RA et al. (2014) "Requirements for pseudosubstrate arginine residues during autoinhibition and phosphatidylinositol 3,4,5-(PO₄)₃-dependent activation of atypical PKC", The Journal of biological chemistry, 289, 25021-30
- Huang X et al. (2003) "Crystal structure of an inactive Akt2 kinase domain", Structure (London, England : 1993), 11, 21-30
- Truebestein L et al. (2021) "Structure of autoinhibited Akt1 reveals mechanism of PIP(3)-mediated activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Lučić I et al. (2018) "Conformational sampling of membranes by Akt controls its activation and inactivation", Proceedings of the National Academy of Sciences of the United States of America, 115, E3940-E3949
Autoinhibited structure

Activated structure

11 structures for P41743
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1VD2 | NMR | - | A | 25-108 | PDB |
| 1WMH | X-ray | 150 A | A | 25-108 | PDB |
| 1ZRZ | X-ray | 300 A | A | 233-596 | PDB |
| 3A8W | X-ray | 210 A | A/B | 249-588 | PDB |
| 3A8X | X-ray | 200 A | A/B | 249-588 | PDB |
| 3ZH8 | X-ray | 274 A | A/B/C | 248-594 | PDB |
| 5LI1 | X-ray | 200 A | A | 248-596 | PDB |
| 5LI9 | X-ray | 179 A | A | 248-596 | PDB |
| 5LIH | X-ray | 325 A | A/B | 248-596 | PDB |
| 6ILZ | X-ray | 326 A | A/C/E/G | 249-588 | PDB |
| AF-P41743-F1 | Predicted | AlphaFoldDB |
312 variants for P41743
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA355095851 rs1305218534 |
2 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA355095849 rs1305218534 |
2 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1082973 CA87676314 |
4 | Q>H | No |
ClinGen gnomAD |
|
|
CA355095884 rs1238949875 |
5 | R>W | No |
ClinGen gnomAD |
|
|
CA355095901 rs1309785216 |
6 | D>G | No |
ClinGen gnomAD |
|
|
CA355095895 rs1444111261 |
6 | D>N | No |
ClinGen TOPMed |
|
|
rs746473761 CA2700305 |
7 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1194363431 CA355095934 |
8 | S>R | No |
ClinGen gnomAD |
|
|
CA87676329 rs979336423 |
9 | T>N | No |
ClinGen Ensembl |
|
|
rs146517821 CA2700308 |
10 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700307 rs781622043 |
10 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1215675569 CA355095969 |
11 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1180800394 CA355095978 |
12 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA87676347 rs935282042 |
13 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs770401419 CA2700309 |
14 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA355096024 rs1179487149 |
16 | G>C | No |
ClinGen gnomAD |
|
|
rs763427114 CA2700312 |
16 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1577333820 CA355096030 |
17 | G>S | No |
ClinGen Ensembl |
|
|
CA355096045 rs1453580506 |
18 | G>C | No |
ClinGen TOPMed gnomAD |
|
|
CA2700316 rs554225297 |
19 | S>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA355096053 rs1182925435 |
19 | S>R | No |
ClinGen TOPMed |
|
|
CA2700317 rs572493199 |
21 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1397484473 CA355096092 |
22 | H>Y | No |
ClinGen gnomAD |
|
|
CA355096120 rs1445671164 |
24 | H>Y | No |
ClinGen gnomAD |
|
|
rs1331532531 CA355096154 |
26 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1337327825 CA355096167 |
27 | R>Q | No |
ClinGen gnomAD |
|
|
rs1283562436 CA355096207 |
30 | A>V | No |
ClinGen gnomAD |
|
|
CA2700319 rs765259096 |
31 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355096237 rs1256310680 |
33 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA355096239 rs1256310680 |
33 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1457319386 CA355096243 |
33 | R>H | No |
ClinGen gnomAD |
|
|
CA355096254 rs1252865018 |
34 | G>A | No |
ClinGen gnomAD |
|
|
CA355096251 rs1199520604 |
34 | G>W | No |
ClinGen gnomAD |
|
|
CA355098398 rs1292998335 |
39 | T>I | No |
ClinGen TOPMed |
|
|
CA2700342 rs751420634 |
40 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs751420634 CA2700343 |
40 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs766148364 CA2700341 |
40 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs961288557 CA87658270 |
43 | P>T | No |
ClinGen TOPMed |
|
|
rs202068509 CA87658273 |
50 | L>R | No |
ClinGen Ensembl |
|
|
rs1296298304 CA355098478 |
51 | C>W | No |
ClinGen gnomAD |
|
|
CA2700344 rs767204074 |
52 | N>H | No |
ClinGen ExAC gnomAD |
|
|
CA2700345 rs369333148 |
52 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700346 rs755677529 |
55 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs1280748335 CA355098512 |
57 | M>L | No |
ClinGen TOPMed |
|
|
rs1232817418 CA355098517 |
57 | M>T | No |
ClinGen gnomAD |
|
|
CA87658291 rs1032101463 |
58 | C>Y | No |
ClinGen Ensembl |
|
|
CA355098535 rs1256752939 |
60 | F>I | No |
ClinGen gnomAD |
|
|
CA355098549 rs1403982825 |
61 | D>E | No |
ClinGen TOPMed |
|
|
CA355098547 rs1350537092 |
61 | D>G | No |
ClinGen gnomAD |
|
|
rs140807328 CA2700349 |
62 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1272447670 CA355098555 |
62 | N>S | No |
ClinGen gnomAD |
|
|
rs757899632 CA2700350 |
63 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2700351 rs757899632 |
63 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2700353 rs772402293 |
64 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA2700354 rs775805215 |
65 | L>R | No |
ClinGen ExAC gnomAD |
|
|
rs201089076 CA87658313 |
66 | F>L | No |
ClinGen 1000Genomes |
|
|
CA355098578 rs1197750201 |
66 | F>Y | No |
ClinGen gnomAD |
|
|
CA2700356 rs768794063 |
67 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs776632465 CA2700357 |
71 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs762941664 CA2700358 |
73 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA355098645 COSM582652 rs1478749679 |
75 | G>* | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1321696346 CA355099342 |
75 | G>E | No |
ClinGen gnomAD |
|
|
rs760374153 CA355099352 |
76 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA2700384 rs763696294 |
77 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA2700388 rs750971070 |
86 | L>* | No |
ClinGen ExAC gnomAD |
|
|
CA87669294 rs370916263 |
87 | E>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370916263 CA2700390 |
87 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700389 rs182924386 |
87 | E>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA87669307 rs982119706 |
89 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA355099432 rs982119706 |
89 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1170271274 CA355099455 |
92 | L>P | No |
ClinGen gnomAD |
|
|
rs902957582 CA87669310 |
92 | L>V | No |
ClinGen Ensembl |
|
|
CA87669320 rs111567328 |
95 | L>P | No |
ClinGen Ensembl |
|
|
rs1444607173 CA355099497 |
98 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA355099498 rs1444607173 |
98 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
CA2700393 rs781289139 |
99 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA2700394 rs748182793 |
102 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355099542 rs1271977652 |
105 | V>L | No |
ClinGen TOPMed |
|
|
CA355099784 rs1214977029 |
107 | P>A | No |
ClinGen gnomAD |
|
|
rs768340835 CA2700418 |
109 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA355099797 rs1202631310 |
109 | V>I | No |
ClinGen TOPMed |
|
|
rs368704903 COSM1291649 CA2700423 |
112 | R>C | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs980860031 CA87672265 COSM208049 |
112 | R>H | large_intestine endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA2700422 rs368704903 |
112 | R>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs762390072 CA2700424 |
115 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1224423994 CA355099832 |
115 | M>V | No |
ClinGen TOPMed |
|
|
CA355099849 rs1379196093 |
117 | C>S | No |
ClinGen TOPMed |
|
|
rs766866155 CA2700425 |
121 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA355099905 rs1279890266 |
123 | S>C | No |
ClinGen gnomAD |
|
|
CA87676413 rs946756475 |
124 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs567380681 CA2700443 |
126 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs567380681 CA87676416 |
126 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs146841636 CA87676426 |
127 | R>K | No |
ClinGen Ensembl |
|
|
rs56154494 CA2700445 VAR_042323 |
130 | R>C | No |
ClinGen UniProt ExAC TOPMed dbSNP gnomAD |
|
|
CA2700446 rs369872734 |
130 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs775812965 CA355099950 |
131 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs775812965 CA2700448 |
131 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs1187831601 CA355100000 |
138 | A>T | No |
ClinGen TOPMed |
|
|
rs373086682 CA2700450 |
139 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA355100031 rs1204274368 |
142 | T>S | No |
ClinGen TOPMed |
|
|
CA2700453 rs765219909 |
146 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA87676473 rs762292026 |
147 | R>H | No |
ClinGen TOPMed |
|
|
rs750420887 CA2700472 |
151 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763154483 CA2700473 |
155 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs752691470 CA2700475 |
158 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs1293622637 CA355100166 |
160 | R>Q | No |
ClinGen TOPMed |
|
|
rs1289447991 CA355100193 |
164 | L>F | No |
ClinGen gnomAD |
|
|
CA355100200 rs1361108822 |
165 | G>E | No |
ClinGen gnomAD |
|
|
CA87678592 rs1030581930 |
166 | R>H | No |
ClinGen TOPMed |
|
|
rs1259133115 CA355100211 |
167 | Q>R | No |
ClinGen gnomAD |
|
|
rs185526558 CA87678602 |
169 | Y>C | No |
ClinGen 1000Genomes |
|
|
CA87678596 rs1050315708 |
169 | Y>H | No |
ClinGen TOPMed |
|
|
rs148843944 CA355100249 |
172 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2700479 rs756841596 |
172 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA2700482 rs771576001 |
173 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355100275 rs1436578624 |
176 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1436578624 CA355100274 |
176 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA87678724 rs750524485 |
178 | V>I | No |
ClinGen Ensembl |
|
|
rs1382818070 CA355100328 |
183 | H>R | No |
ClinGen gnomAD |
|
|
rs868417022 CA87678730 |
185 | L>I | No |
ClinGen Ensembl |
|
|
CA355100344 COSM3945125 rs1329586167 |
186 | V>I | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA2700486 rs776837368 |
187 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs147562215 CA2700488 |
188 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2700487 rs376895151 |
188 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA355100367 rs1238680908 |
189 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
CA355100377 rs1260622724 |
191 | G>R | No |
ClinGen gnomAD |
|
|
CA2700490 rs763066411 |
192 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2700489 COSM1222039 rs141965722 |
192 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA2700492 rs766551522 |
197 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355100416 rs1192616664 |
197 | Q>R | No |
ClinGen gnomAD |
|
|
CA2700518 rs774546861 |
200 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs111317464 CA87680754 |
201 | M>V | No |
ClinGen Ensembl |
|
|
rs763902996 CA355100467 |
203 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763902996 CA2700521 |
203 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs71277168 CA2700522 |
204 | D>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2700524 rs374000097 |
205 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs750284734 CA2700525 |
208 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs750284734 CA87680780 |
208 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA2700526 rs758015497 |
209 | H>N | No |
ClinGen ExAC gnomAD |
|
|
CA355100551 rs766040399 |
215 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs763072369 CA355100554 |
215 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA87680798 rs763072369 |
215 | T>R | No |
ClinGen TOPMed gnomAD |
|
|
rs766040399 CA2700527 |
215 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs767045797 CA355100575 |
217 | I>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1435957709 CA355100579 |
217 | I>M | No |
ClinGen gnomAD |
|
|
rs767045797 CA2700557 |
217 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2700559 rs191938996 |
219 | Y>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2700560 rs370513574 |
222 | S>* | No |
ClinGen ESP ExAC |
|
|
rs757703152 CA2700562 |
224 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2700561 rs754218167 |
224 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756263490 CA87682570 |
227 | L>S | No |
ClinGen Ensembl |
|
|
rs147104137 CA2700564 |
229 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA355100674 rs1301070129 |
231 | G>V | No |
ClinGen gnomAD |
|
|
CA2700565 rs772250699 |
234 | K>T | No |
ClinGen ExAC gnomAD |
|
|
rs199997573 CA2700583 |
236 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355100728 rs1441299379 |
237 | M>I | No |
ClinGen TOPMed |
|
|
rs747042200 CA2700584 |
237 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768734145 CA2700585 |
239 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA355100756 rs1560181245 |
241 | E>G | No |
ClinGen Ensembl |
|
|
CA355100767 rs1178271455 |
243 | G>C | No |
ClinGen gnomAD |
|
|
rs200726509 CA2700587 |
243 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1458333589 CA355100797 |
247 | S>F | No |
ClinGen gnomAD |
|
|
CA355100836 rs1393309823 |
253 | D>G | No |
ClinGen gnomAD |
|
|
CA2700589 rs774168806 |
255 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774168806 CA355100848 |
255 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771711948 CA2700591 |
258 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs138560118 CA2700590 |
258 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs879138314 CA87686337 |
271 | V>F | No |
ClinGen Ensembl |
|
|
CA355100956 rs1335738205 |
272 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
rs762380206 CA2700597 |
272 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2700600 rs150239950 |
273 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700601 rs750752062 |
274 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA355100979 rs1402498314 |
275 | K>N | No |
ClinGen Ensembl |
|
|
CA87686401 rs867140849 |
276 | T>A | No |
ClinGen Ensembl |
|
|
CA355100995 COSM284260 rs1428010723 |
278 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1254638218 CA355100998 |
278 | R>H | No |
ClinGen gnomAD |
|
|
CA2700603 rs368607450 |
284 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs766563408 CA2700604 |
285 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755258716 CA2700606 |
289 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs973528502 CA87686466 |
291 | N>D | No |
ClinGen Ensembl |
|
|
rs1329248941 CA355101093 |
292 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA2700608 rs748272532 |
293 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA2700629 rs573160898 |
295 | D>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA355101134 rs1425394515 |
296 | I>V | No |
ClinGen TOPMed |
|
|
rs1228343200 CA355101143 |
297 | D>G | No |
ClinGen gnomAD |
|
|
rs752731655 CA2700630 |
298 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA355101161 rs1281629002 |
300 | Q>* | No |
ClinGen gnomAD |
|
|
CA355101180 rs1379694217 |
302 | E>G | No |
ClinGen TOPMed |
|
|
rs1234702783 CA355101223 |
308 | Q>E | No |
ClinGen TOPMed |
|
|
CA2700632 rs778855961 |
308 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs745782136 CA2700633 |
310 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs977008114 CA87687050 |
312 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA2700635 rs779686819 |
319 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1172744102 CA355101352 |
327 | R>G | No |
ClinGen gnomAD |
|
|
rs777394724 CA355101374 |
328 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA87687542 rs950321169 |
329 | F>L | No |
ClinGen TOPMed |
|
|
CA355101401 rs1232048013 |
332 | I>T | No |
ClinGen gnomAD |
|
|
CA355101398 rs1370173894 |
332 | I>V | No |
ClinGen gnomAD |
|
|
rs980725247 CA87687565 |
337 | G>R | No |
ClinGen TOPMed |
|
|
rs1560181937 CA355101525 |
349 | K>I | No |
ClinGen Ensembl |
|
|
CA355101557 rs1399769269 |
354 | H>D | No |
ClinGen TOPMed |
|
|
CA355101573 rs1386397434 |
356 | R>T | No |
ClinGen TOPMed |
|
|
rs927803592 CA87689386 |
358 | Y>H | No |
ClinGen TOPMed |
|
|
CA2700678 rs752586080 |
359 | S>A | No |
ClinGen ExAC gnomAD |
|
|
COSM125590 CA2700679 rs554687623 |
359 | S>C | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC gnomAD |
|
CA355101634 rs1224995597 |
363 | S>G | No |
ClinGen gnomAD |
|
|
CA2700680 rs200664120 |
364 | L>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs770485461 CA2700682 |
365 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778456196 CA2700683 |
369 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs746373616 CA2700684 |
372 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs772611163 CA2700685 |
372 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2700687 rs761124312 |
376 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA355101771 rs1373137084 |
383 | N>S | No |
ClinGen gnomAD |
|
|
CA87689421 rs201380049 |
392 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs773463648 COSM1222038 CA2700694 |
398 | G>S | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA355101894 rs1230577514 |
401 | K>E | No |
ClinGen gnomAD |
|
|
CA87695170 rs1008561303 |
405 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1008561303 CA355102070 |
405 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA2700714 rs763152419 |
405 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1332841310 CA355102074 |
406 | P>A | No |
ClinGen TOPMed |
|
|
rs374177981 CA2700716 |
406 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374177981 CA2700717 |
406 | P>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1193411428 CA355102095 |
409 | T>R | No |
ClinGen gnomAD |
|
|
rs900136662 CA87695213 |
410 | T>A | No |
ClinGen TOPMed |
|
|
rs1560185295 CA355102102 |
411 | S>G | No |
ClinGen Ensembl |
|
|
rs1560185295 CA355102101 |
411 | S>R | No |
ClinGen Ensembl |
|
|
rs763947334 CA2700718 |
415 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs1560185302 CA355102143 |
417 | P>A | No |
ClinGen Ensembl |
|
|
CA355102205 rs1182254154 |
426 | R>G | No |
ClinGen TOPMed |
|
|
rs756884894 CA2700720 |
426 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA355102226 rs1458431350 |
429 | D>H | No |
ClinGen gnomAD |
|
|
CA2700745 rs773224124 |
431 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2700746 rs773224124 COSM4149659 |
431 | G>V | ovary [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs201740893 CA87696503 |
432 | F>L | No |
ClinGen 1000Genomes |
|
|
rs1483055491 CA355102263 |
432 | F>L | No |
ClinGen gnomAD |
|
|
rs374883456 CA2700748 |
439 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700750 rs184216798 |
443 | M>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2700751 rs779029814 |
446 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs1364845286 CA355102358 |
446 | M>V | No |
ClinGen TOPMed |
|
|
rs745958617 CA2700752 |
447 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1388083127 CA355102385 |
450 | R>G | No |
ClinGen gnomAD |
|
|
rs1577375356 CA355102394 |
451 | S>A | No |
ClinGen Ensembl |
|
|
rs1161973892 CA355102416 |
454 | D>G | No |
ClinGen TOPMed |
|
|
rs768597939 CA2700753 |
455 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776558522 CA2700754 |
456 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355102428 rs1372532118 |
456 | V>I | No |
ClinGen gnomAD |
|
|
CA355102455 rs1450462772 |
460 | D>G | No |
ClinGen gnomAD |
|
|
rs1341309666 CA355102451 |
460 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1341870333 CA355102480 |
463 | D>E | No |
ClinGen gnomAD |
|
|
rs554559908 CA2700758 |
464 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs765967034 CA2700759 |
466 | T>I | No |
ClinGen ExAC |
|
|
rs751036713 CA2700760 |
470 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1485937829 CA355102534 |
471 | F>S | No |
ClinGen TOPMed |
|
|
rs1378322375 CA355098661 |
474 | I>F | No |
ClinGen gnomAD |
|
|
rs761243863 CA2700782 |
475 | L>S | No |
ClinGen ExAC gnomAD |
|
|
rs1388201066 COSM78349 CA355098706 |
480 | R>C | ovary pancreas large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs754216126 CA2700785 |
483 | R>C | No |
ClinGen ExAC gnomAD |
|
|
COSM1420563 CA355098725 rs1389006052 |
483 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA87662365 rs17855494 |
485 | L>M | No |
ClinGen Ensembl |
|
|
rs1302542745 CA355098771 |
491 | S>G | No |
ClinGen gnomAD |
|
|
CA2700787 rs757566946 |
493 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA87663224 rs373682853 |
500 | D>E | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1577377456 CA355098858 |
501 | P>L | No |
ClinGen Ensembl |
|
|
rs1469931815 CA355098855 |
501 | P>S | No |
ClinGen gnomAD |
|
|
rs762444412 CA2700808 |
504 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs17856646 CA87663245 |
508 | H>L | No |
ClinGen Ensembl |
|
|
rs1319504292 CA355098949 |
515 | D>N | No |
ClinGen gnomAD |
|
|
rs974333981 CA87663249 |
516 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA355098956 rs1282299429 |
516 | I>V | No |
ClinGen TOPMed |
|
|
CA355098975 rs1577377491 |
519 | H>N | No |
ClinGen Ensembl |
|
|
rs770998502 CA2700811 |
520 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA87663269 rs866058843 |
523 | R>* | No |
ClinGen gnomAD |
|
|
rs368665283 CA87663273 COSM1041063 |
523 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP gnomAD |
|
CA355099038 rs1423404616 |
528 | D>N | No |
ClinGen gnomAD |
|
|
CA355099043 rs1165933429 |
528 | D>V | No |
ClinGen gnomAD |
|
|
CA355099051 rs1560187209 |
529 | M>I | No |
ClinGen Ensembl |
|
|
CA2700815 rs781306402 |
529 | M>V | No |
ClinGen ExAC |
|
|
CA2700834 rs752869987 |
530 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA355099081 rs1296524027 |
531 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs199683302 CA87664538 |
532 | Q>R | No |
ClinGen Ensembl |
|
|
CA355099097 rs1330922659 |
534 | Q>* | No |
ClinGen TOPMed |
|
|
CA2700836 rs142234162 |
535 | V>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2700835 rs142234162 |
535 | V>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA355099117 rs1216250049 |
537 | P>S | No |
ClinGen gnomAD |
|
|
rs750120400 CA2700837 |
539 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355099141 rs1311650226 |
540 | K>N | No |
ClinGen gnomAD |
|
|
CA2700838 rs369277385 |
542 | N>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs780053668 CA2700839 |
542 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs780053668 CA2700840 |
542 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA2700841 rs768466580 |
543 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355099166 rs1208504932 |
545 | G>R | No |
ClinGen gnomAD |
|
|
CA355099170 rs1458271182 |
545 | G>V | No |
ClinGen TOPMed |
|
|
rs1249036295 CA355099191 |
548 | G>D | No |
ClinGen gnomAD |
|
|
rs1189963071 CA355099229 |
553 | D>G | No |
ClinGen gnomAD |
|
|
rs1414651390 CA355099279 |
560 | P>A | No |
ClinGen TOPMed |
|
|
rs17851954 CA2700844 |
560 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs17851954 CA87664624 |
560 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs139167704 CA2700846 |
563 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700847 rs771277289 |
565 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA355099326 rs371799261 |
567 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs145084763 CA2700849 |
568 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2700869 rs777211672 |
570 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA355099572 rs1305100978 |
571 | V>M | No |
ClinGen gnomAD |
|
|
rs749763295 CA2700870 |
572 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs1228061878 CA355099588 |
573 | K>R | No |
ClinGen gnomAD |
|
|
CA355099596 rs1280082949 |
574 | I>T | No |
ClinGen TOPMed |
|
|
rs1353052159 CA355099623 |
578 | E>* | No |
ClinGen gnomAD |
|
|
rs1475798615 CA355099650 |
581 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
CA2700871 rs374370739 |
585 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA355099685 rs1225351390 |
586 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA355099689 rs1309853880 |
587 | P>T | No |
ClinGen TOPMed |
|
|
CA2700875 rs775797042 |
594 | E>K | No |
ClinGen ExAC TOPMed |
No associated diseases with P41743
13 regional properties for P41743
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | PB1 domain | 25 - 108 | IPR000270 |
| domain | Protein kinase domain | 254 - 522 | IPR000719 |
| domain | AGC-kinase, C-terminal | 523 - 594 | IPR000961 |
| domain | Protein kinase C-like, phorbol ester/diacylglycerol-binding domain | 140 - 192 | IPR002219 |
| active_site | Serine/threonine-protein kinase, active site | 374 - 386 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 260 - 287 | IPR017441 |
| domain | Protein kinase, C-terminal | 551 - 584 | IPR017892 |
| domain | Diacylglycerol/phorbol-ester binding | 138 - 152 | IPR020454-1 |
| domain | Diacylglycerol/phorbol-ester binding | 154 - 163 | IPR020454-2 |
| domain | Diacylglycerol/phorbol-ester binding | 167 - 178 | IPR020454-3 |
| domain | Diacylglycerol/phorbol-ester binding | 179 - 191 | IPR020454-4 |
| domain | Atypical protein kinase C iota type, catalytic domain | 233 - 596 | IPR034661 |
| domain | Protein kinase C, PB1 domain | 26 - 108 | IPR034877 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24351 | RIBOSOMAL PROTEIN S6 KINASE |
| PANTHER Subfamily | PTHR24351:SF236 | PROTEIN KINASE C IOTA TYPE |
| PANTHER Protein Class | protein modifying enzyme | |
| PANTHER Pathway Category | No pathway information available | |
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| bicellular tight junction | An occluding cell-cell junction that is composed of a branching network of sealing strands that completely encircles the apical end of each cell in an epithelial sheet; the outer leaflets of the two interacting plasma membranes are seen to be tightly apposed where sealing strands are present. Each sealing strand is composed of a long row of transmembrane adhesion proteins embedded in each of the two interacting plasma membranes. |
| cell leading edge | The area of a motile cell closest to the direction of movement. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| intercellular bridge | A direct connection between the cytoplasm of two cells that is formed following the completion of cleavage furrow ingression during cell division. They are usually present only briefly prior to completion of cytokinesis. However, in some cases, such as the bridges between germ cells during their development, they become stabilised. |
| microtubule cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| PAR polarity complex | A protein kinase complex that is required for the establishment of a cell polarity axis during the cell division cycle. Binds directly to activated CDC42 GTPase and is required for orchestrating a cellular gradient of CDC42. In S. cerevisiae components are: BEM1, CDC24 and CLA4; from worms to vertebrates it contains a PAR6 protein, PAR3 protein and an atypical PKC. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| Schaffer collateral - CA1 synapse | A synapse between the Schaffer collateral axon of a CA3 pyramidal cell and a CA1 pyramidal cell. |
| Schmidt-Lanterman incisure | Regions within compact myelin in which the cytoplasmic faces of the enveloping myelin sheath are not tightly juxtaposed, and include cytoplasm from the cell responsible for making the myelin. Schmidt-Lanterman incisures occur in the compact myelin internode, while lateral loops are analogous structures found in the paranodal region adjacent to the nodes of Ranvier. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein kinase C activity | Calcium-dependent catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. |
| metal ion binding | Binding to a metal ion. |
| phospholipid binding | Binding to a phospholipid, a class of lipids containing phosphoric acid as a mono- or diester. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein kinase C activity | Catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. This reaction requires diacylglycerol. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
28 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cell-cell junction organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cell-cell junction. A cell-cell junction is a specialized region of connection between two cells. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures. |
| establishment of apical/basal cell polarity | The specification and formation of the polarity of a cell along its apical/basal axis. |
| establishment or maintenance of epithelial cell apical/basal polarity | Any cellular process that results in the specification, formation or maintenance of the apicobasal polarity of an epithelial cell. |
| eye photoreceptor cell development | Development of a photoreceptor, a sensory cell in the eye that reacts to the presence of light. They usually contain a pigment that undergoes a chemical change when light is absorbed, thus stimulating a nerve. |
| Golgi vesicle budding | The evagination of the Golgi membrane, resulting in formation of a vesicle. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| membrane organization | A process which results in the assembly, arrangement of constituent parts, or disassembly of a membrane. A membrane is a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of glial cell apoptotic process | Any process that stops, prevents, or reduces the frequency, rate, or extent of glial cell apoptotic process. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of endothelial cell apoptotic process | Any process that activates or increases the frequency, rate or extent of endothelial cell apoptotic process. |
| positive regulation of glial cell proliferation | Any process that activates or increases the rate or extent of glial cell proliferation. |
| positive regulation of glucose import | Any process that activates or increases the frequency, rate or extent of the import of the hexose monosaccharide glucose into a cell or organelle. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of NF-kappaB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB. |
| positive regulation of Notch signaling pathway | Any process that activates or increases the frequency, rate or extent of the Notch signaling pathway. |
| positive regulation of protein localization to plasma membrane | Any process that activates or increases the frequency, rate or extent of protein localization to plasma membrane. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| protein targeting to membrane | The process of directing proteins towards a membrane, usually using signals contained within the protein. |
| regulation of postsynaptic membrane neurotransmitter receptor levels | Any process that regulates the the local concentration of neurotransmitter receptor at the postsynaptic membrane. |
| response to interleukin-1 | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| secretion | The controlled release of a substance by a cell or a tissue. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
30 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P24583 | PKC1 | Protein kinase C-like 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| A1Z9X0 | aPKC | Atypical protein kinase C | Drosophila melanogaster (Fruit fly) | SS |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens (Human) | EV |
| Q05513 | PRKCZ | Protein kinase C zeta type | Homo sapiens (Human) | SS |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| Q96BR1 | SGK3 | Serine/threonine-protein kinase Sgk3 | Homo sapiens (Human) | SS |
| Q9HBY8 | SGK2 | Serine/threonine-protein kinase Sgk2 | Homo sapiens (Human) | SS |
| O00141 | SGK1 | Serine/threonine-protein kinase Sgk1 | Homo sapiens (Human) | PR |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | Homo sapiens (Human) | EV |
| Q9Y2H1 | STK38L | Serine/threonine-protein kinase 38-like | Homo sapiens (Human) | EV |
| Q6A1A2 | PDPK2P | Putative 3-phosphoinositide-dependent protein kinase 2 | Homo sapiens (Human) | PR |
| Q02956 | Prkcz | Protein kinase C zeta type | Mus musculus (Mouse) | SS |
| Q62074 | Prkci | Protein kinase C iota type | Mus musculus (Mouse) | SS |
| P09217 | Prkcz | Protein kinase C zeta type | Rattus norvegicus (Rat) | SS |
| F1M7Y5 | Prkci | Protein kinase C iota type | Rattus norvegicus (Rat) | SS |
| Q19266 | pkc-3 | Protein kinase C-like 3 | Caenorhabditis elegans | SS |
| Q9SUA3 | D6PKL1 | Serine/threonine-protein kinase D6PKL1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q90XF2 | prkci | Protein kinase C iota type | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPTQRDSSTM | SHTVAGGGSG | DHSHQVRVKA | YYRGDIMITH | FEPSISFEGL | CNEVRDMCSF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DNEQLFTMKW | IDEEGDPCTV | SSQLELEEAF | RLYELNKDSE | LLIHVFPCVP | ERPGMPCPGE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DKSIYRRGAR | RWRKLYCANG | HTFQAKRFNR | RAHCAICTDR | IWGLGRQGYK | CINCKLLVHK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KCHKLVTIEC | GRHSLPQEPV | MPMDQSSMHS | DHAQTVIPYN | PSSHESLDQV | GEEKEAMNTR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESGKASSSLG | LQDFDLLRVI | GRGSYAKVLL | VRLKKTDRIY | AMKVVKKELV | NDDEDIDWVQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TEKHVFEQAS | NHPFLVGLHS | CFQTESRLFF | VIEYVNGGDL | MFHMQRQRKL | PEEHARFYSA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EISLALNYLH | ERGIIYRDLK | LDNVLLDSEG | HIKLTDYGMC | KEGLRPGDTT | STFCGTPNYI |
| 430 | 440 | 450 | 460 | 470 | 480 |
| APEILRGEDY | GFSVDWWALG | VLMFEMMAGR | SPFDIVGSSD | NPDQNTEDYL | FQVILEKQIR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IPRSLSVKAA | SVLKSFLNKD | PKERLGCHPQ | TGFADIQGHP | FFRNVDWDMM | EQKQVVPPFK |
| 550 | 560 | 570 | 580 | 590 | |
| PNISGEFGLD | NFDSQFTNEP | VQLTPDDDDI | VRKIDQSEFE | GFEYINPLLM | SAEECV |