P41377
Gene name |
TIF4A-2 (RH19, At1g54270, F20D21.9, F20D21_52) |
Protein name |
Eukaryotic initiation factor 4A-2 |
Names |
eIF-4A-2, ATP-dependent RNA helicase eIF4A-2, DEAD-box ATP-dependent RNA helicase 19 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G54270 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
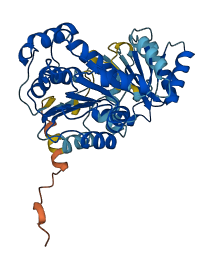
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P41377
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P41377-F1 | Predicted | AlphaFoldDB |
6 variants for P41377
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_1_20260520_C_A | 9 | T>K | No | 1000Genomes | |
| tmp_1_20260708_C_A | 43 | A>D | No | 1000Genomes | |
| ENSVATH14314307 | 178 | R>H | No | 1000Genomes | |
| ENSVATH14314308 | 207 | Q>R | No | 1000Genomes | |
| tmp_1_20261410_C_T | 211 | P>S | No | 1000Genomes | |
| tmp_1_20261450_C_T | 224 | P>L | No | 1000Genomes |
No associated diseases with P41377
5 regional properties for P41377
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | ATP-dependent RNA helicase DEAD-box, conserved site | 186 - 194 | IPR000629 |
| domain | Helicase, C-terminal | 251 - 412 | IPR001650 |
| domain | DEAD/DEAH box helicase domain | 64 - 225 | IPR011545 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 58 - 255 | IPR014001 |
| domain | RNA helicase, DEAD-box type, Q motif | 39 - 67 | IPR014014 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plant-type vacuole | A closed structure that is completely surrounded by a unit membrane, contains liquid, and retains the same shape regardless of cell cycle phase. An example of this structure is found in Arabidopsis thaliana. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| plasmodesma | A fine cytoplasmic channel, found in all higher plants, that connects the cytoplasm of one cell to that of an adjacent cell. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| mRNA binding | Binding to messenger RNA (mRNA), an intermediate molecule between DNA and protein. mRNA includes UTR and coding sequences, but does not contain introns. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
| translation initiation factor activity | Functions in the initiation of ribosome-mediated translation of mRNA into a polypeptide. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| cytoplasmic translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein in the cytoplasm. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P41378 | Eukaryotic initiation factor 4A | Triticum aestivum (Wheat) | PR | |
| P10081 | TIF2 | ATP-dependent RNA helicase eIF4A | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q3SZ54 | EIF4A1 | Eukaryotic initiation factor 4A-I | Bos taurus (Bovine) | PR |
| Q3SZ65 | EIF4A2 | Eukaryotic initiation factor 4A-II | Bos taurus (Bovine) | PR |
| Q8JFP1 | EIF4A2 | Eukaryotic initiation factor 4A-II | Gallus gallus (Chicken) | PR |
| A5A6N4 | EIF4A1 | Eukaryotic initiation factor 4A-I | Pan troglodytes (Chimpanzee) | PR |
| Q14240 | EIF4A2 | Eukaryotic initiation factor 4A-II | Homo sapiens (Human) | PR |
| P60842 | EIF4A1 | Eukaryotic initiation factor 4A-I | Homo sapiens (Human) | PR |
| Q41741 | Eukaryotic initiation factor 4A | Zea mays (Maize) | PR | |
| P60843 | Eif4a1 | Eukaryotic initiation factor 4A-I | Mus musculus (Mouse) | PR |
| P10630 | Eif4a2 | Eukaryotic initiation factor 4A-II | Mus musculus (Mouse) | PR |
| Q5RKI1 | Eif4a2 | Eukaryotic initiation factor 4A-II | Rattus norvegicus (Rat) | PR |
| P35683 | Os06g0701100 | Eukaryotic initiation factor 4A-1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z2Z4 | Os02g0146600 | Eukaryotic initiation factor 4A-3 | Oryza sativa subsp japonica (Rice) | PR |
| P27639 | inf-1 | Eukaryotic initiation factor 4A | Caenorhabditis elegans | PR |
| Q3E9C3 | RH58 | DEAD-box ATP-dependent RNA helicase 58, chloroplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9CAI7 | TIF4A-3 | Eukaryotic initiation factor 4A-3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SA27 | RH36 | DEAD-box ATP-dependent RNA helicase 36 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FVV4 | RH55 | DEAD-box ATP-dependent RNA helicase 55 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O80792 | RH33 | Putative DEAD-box ATP-dependent RNA helicase 33 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22907 | RH24 | DEAD-box ATP-dependent RNA helicase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIH9 | RH51 | DEAD-box ATP-dependent RNA helicase 51 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q56X76 | RH39 | DEAD-box ATP-dependent RNA helicase 39 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GY84 | RH10 | DEAD-box ATP-dependent RNA helicase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FFQ1 | RH31 | DEAD-box ATP-dependent RNA helicase 31 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SB89 | RH27 | DEAD-box ATP-dependent RNA helicase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41376 | EIF4A1 | Eukaryotic initiation factor 4A-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84TG1 | RH57 | DEAD-box ATP-dependent RNA helicase 57 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8H0U8 | RH42 | DEAD-box ATP-dependent RNA helicase 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGSAPEGTQ | FDTRQFDQRL | NEVLDGQDEF | FTSYDEVHES | FDAMGLQENL | LRGIYAYGFE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KPSAIQQRGI | VPFCKGLDVI | QQAQSGTGKT | ATFCSGVLQQ | LDYALLQCQA | LVLAPTRELA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QQIEKVMRAL | GDYQGVKVHA | CVGGTSVRED | QRILQAGVHV | VVGTPGRVFD | MLRRQSLRPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| CIKMFVLDEA | DEMLSRGFKD | QIYDIFQLLP | PKIQVGVFSA | TMPPEALEIT | RKFMSKPVRI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LVKRDELTLE | GIKQFYVNVE | KEDWKLETLC | DLYETLAITQ | SVIFVNTRRK | VDWLTDKMRS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RDHTVSATHG | DMDQNTRDII | MREFRSGSSR | VLITTDLLAR | GIDVQQVSLV | INFDLPTQPE |
| 370 | 380 | 390 | 400 | 410 | |
| NYLHRIGRSG | RFGRKGVAIN | FVTLDDQRML | FDIQKFYNVV | VEELPSNVAD | LL |