P41161
Gene name |
ETV5 (ERM) |
Protein name |
ETS translocation variant 5 |
Names |
Ets-related protein ERM |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2119 |
EC number |
|
Protein Class |
|
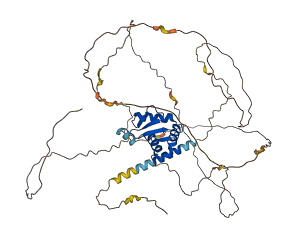
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
367-452 (ETS domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for P41161
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4UNO | X-ray | 195 A | A | 365-462 | PDB |
| 5ILV | X-ray | 180 A | A | 366-457 | PDB |
| AF-P41161-F1 | Predicted | AlphaFoldDB |
320 variants for P41161
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 2 | D>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs771889712 CA2742777 |
3 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA2742776 rs138565774 |
4 | F>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2742775 rs774084292 |
4 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA355845589 rs1410515294 |
6 | D>N | No |
ClinGen TOPMed |
|
|
rs770722736 CA2742774 |
11 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs748760207 CA2742773 |
14 | P>S | No |
ClinGen ExAC |
|
|
rs777457522 CA2742772 |
15 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 15 | G>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2742760 rs767365092 |
16 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs990808893 CA89858749 |
17 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1350949096 CA355845501 |
17 | S>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1189324290 CA355845497 |
18 | R>G | No |
ClinGen TOPMed |
|
|
rs547190842 CA355845494 |
18 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2742759 COSM141070 COSM141069 rs547190842 |
18 | R>Q | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs770669598 CA2742757 |
21 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs763263094 CA2742755 |
25 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742756 rs112315086 |
25 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 28 | I>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355845335 rs1221002517 |
42 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA355845268 rs1433707925 |
49 | D>A | No |
ClinGen gnomAD |
|
|
CA355845258 rs1362660709 |
51 | S>C | No |
ClinGen gnomAD |
|
|
rs1304164243 CA355845255 |
51 | S>N | No |
ClinGen gnomAD |
|
|
rs1358318361 CA355845215 |
56 | A>V | No |
ClinGen gnomAD |
|
|
CA355845167 rs1164415688 |
61 | A>V | No |
ClinGen TOPMed |
|
|
CA355845157 rs1377960309 |
63 | V>L | No |
ClinGen gnomAD |
|
|
CA355845151 rs1411421570 |
64 | P>T | No |
ClinGen gnomAD |
|
|
rs1459088256 CA355845146 |
65 | D>H | No |
ClinGen TOPMed |
|
|
rs1167819091 CA355845141 |
65 | D>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1429227347 CA355845118 |
68 | Q>R | No |
ClinGen gnomAD |
|
|
CA355845110 rs1371359850 |
69 | F>S | No |
ClinGen gnomAD |
|
|
CA355845095 rs1319038497 |
71 | P>L | No |
ClinGen TOPMed |
|
|
CA2742719 rs751536392 |
72 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA2742718 rs142245897 |
74 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1412277860 CA355844729 |
83 | P>A | No |
ClinGen TOPMed |
|
|
rs907103876 CA89856281 |
85 | P>L | No |
ClinGen TOPMed |
|
|
CA89856282 rs540138059 |
85 | P>T | No |
ClinGen gnomAD |
|
|
CA2742684 rs775007271 |
86 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1345723205 CA355844694 |
87 | K>R | No |
ClinGen gnomAD |
|
|
CA355844687 rs1578552734 |
88 | I>V | No |
ClinGen Ensembl |
|
|
CA89856280 rs767971453 |
90 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs767971453 CA2742683 |
90 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1276673113 CA355844668 |
90 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| TCGA novel | 94 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs774972931 CA2742681 |
97 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs771188086 CA2742679 |
98 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773508609 CA2742677 |
100 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs202139916 CA89856279 |
102 | C>Y | No |
ClinGen 1000Genomes |
|
|
rs752940067 CA89856278 |
104 | H>R | No |
ClinGen Ensembl |
|
|
CA2742673 rs755131256 |
106 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA2742672 rs745776119 |
107 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745776119 CA89856277 |
107 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1421479 CA355844548 rs1165631447 COSM1421480 |
107 | A>V | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1249712516 CA355844545 |
108 | L>F | No |
ClinGen TOPMed |
|
|
CA355844538 rs1482734733 |
109 | G>V | No |
ClinGen TOPMed |
|
|
CA355844525 rs1578552686 |
111 | N>S | No |
ClinGen Ensembl |
|
|
rs61760177 CA2742670 |
112 | Y>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs200054486 CA2742671 |
112 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1010586417 CA89856276 |
113 | G>R | No |
ClinGen Ensembl |
|
|
rs557148325 CA89856275 |
114 | E>G | No |
ClinGen 1000Genomes |
|
|
rs763691220 CA2742668 |
116 | C>G | No |
ClinGen ExAC gnomAD |
|
|
rs755799638 CA2742667 |
117 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1205730921 CA355844467 |
120 | Y>D | No |
ClinGen gnomAD |
|
|
rs1272437619 CA355844458 |
121 | C>R | No |
ClinGen gnomAD |
|
|
rs1332934017 CA355844439 |
122 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs377678658 CA2742650 |
124 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2742649 rs752336984 |
125 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA355844396 rs1473039511 |
128 | P>S | No |
ClinGen TOPMed |
|
|
rs780967082 CA2742648 |
129 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs183104300 CA2742646 |
131 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1578552185 CA355844379 |
131 | F>V | No |
ClinGen Ensembl |
|
|
rs754385394 CA2742647 |
131 | F>Y | No |
ClinGen ExAC |
|
|
CA89856184 rs868275534 |
133 | P>S | No |
ClinGen Ensembl |
|
|
CA2742645 rs766752832 |
135 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA2742644 rs763481967 |
136 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs751015849 CA2742643 |
138 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA355844330 rs765682543 |
139 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765682543 CA2742642 |
139 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA89856182 rs971551407 |
140 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs768938900 CA2742639 |
141 | L>F | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 144 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355844307 rs1578552152 |
144 | T>P | No |
ClinGen Ensembl |
|
|
COSM139499 CA2742638 rs760624416 COSM139498 |
148 | P>L | skin [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
RCV000962064 rs78506201 CA2742637 |
149 | L>V | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA2742636 rs772085062 |
150 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA2742635 rs369950562 |
151 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 152 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs537105512 CA2742634 |
152 | P>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA355844243 rs1578552131 |
154 | Q>P | No |
ClinGen Ensembl |
|
|
CA89856180 rs1012984931 |
159 | T>A | No |
ClinGen TOPMed |
|
|
CA2742631 rs780657690 |
159 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355844211 rs1578552111 |
160 | S>P | No |
ClinGen Ensembl |
|
|
rs200984903 CA2742629 |
162 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs865886165 CA89856179 |
163 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 164 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355844185 rs1438063356 |
164 | P>S | No |
ClinGen gnomAD |
|
|
rs1366935891 CA355844171 |
166 | A>G | No |
ClinGen gnomAD |
|
|
rs1560051612 CA355844175 |
166 | A>T | No |
ClinGen Ensembl |
|
|
CA89856178 rs1007785659 |
167 | G>V | No |
ClinGen Ensembl |
|
|
rs779593863 CA2742628 |
168 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355844158 rs1279125978 |
169 | V>L | No |
ClinGen TOPMed |
|
|
CA355844145 rs1343170858 |
171 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA355844124 rs1394401243 |
174 | P>R | No |
ClinGen gnomAD |
|
|
rs1452592219 CA355844121 |
175 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs185636057 CA2742620 |
177 | A>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs185636057 CA2742621 |
177 | A>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs185636057 COSM1251547 COSM1251548 CA2742622 |
177 | A>T | oesophagus large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA89856176 rs1049852538 |
178 | P>L | No |
ClinGen Ensembl |
|
|
rs1240768693 CA355844104 |
178 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1313363913 CA355844100 |
179 | H>D | No |
ClinGen gnomAD |
|
|
CA2742618 rs772183700 |
179 | H>P | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 179 | H>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs759682748 CA355844090 |
180 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1231448078 CA355844093 |
180 | S>P | No |
ClinGen gnomAD |
|
|
rs759682748 CA2742617 |
180 | S>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742615 rs769742567 |
181 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA89856175 rs375514175 |
184 | P>S | No |
ClinGen ESP |
|
|
rs776452133 CA2742614 |
188 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA2742612 rs768381347 COSM160845 |
191 | F>L | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA355844020 rs1374783313 |
191 | F>S | No |
ClinGen gnomAD |
|
|
rs779639231 CA355844014 |
192 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742610 rs779639231 |
192 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1130282 COSM1130281 CA2742611 rs779639231 |
192 | A>V | Variant assessed as Somatic; 0.0 impact. prostate [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 195 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2742606 rs143264159 |
195 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1183980358 CA355843994 |
196 | P>Q | No |
ClinGen gnomAD |
|
|
CA355843983 rs1257471491 |
198 | H>Y | No |
ClinGen gnomAD |
|
|
CA355843939 rs1251428977 |
204 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA355843934 rs1305468737 |
205 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA2742602 rs752963917 |
205 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA355843922 rs1239539716 |
206 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA355843912 rs1228675123 |
207 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA89856174 rs113592076 |
207 | M>V | No |
ClinGen Ensembl |
|
|
CA355843906 rs1311014457 |
208 | P>L | No |
ClinGen gnomAD |
|
|
CA355843909 rs1354537583 |
208 | P>S | No |
ClinGen gnomAD |
|
|
rs199751532 CA89856173 |
211 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
CA2742601 rs767733743 |
211 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA355843823 rs1390926382 |
218 | F>Y | No |
ClinGen gnomAD |
|
|
CA89854590 rs59502066 |
220 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs59502066 CA355843810 |
220 | R>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1578543481 CA355843781 |
224 | E>D | No |
ClinGen Ensembl |
|
|
rs1295340116 CA355843778 |
225 | P>A | No |
ClinGen gnomAD |
|
|
CA355843755 rs1578543462 |
228 | P>S | No |
ClinGen Ensembl |
|
|
rs778535714 CA2742589 |
229 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs972330166 CA89854589 |
230 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA2742588 rs756876054 |
230 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA355843723 rs1206756635 |
233 | P>L | No |
ClinGen TOPMed |
|
|
CA2742587 rs749793277 |
234 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA2742586 rs778213774 |
235 | V>D | No |
ClinGen ExAC gnomAD |
|
|
rs1416810293 CA355843694 |
238 | D>G | No |
ClinGen gnomAD |
|
|
CA2742585 rs756436991 |
239 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA89854588 rs761741466 |
240 | R>C | No |
ClinGen TOPMed |
|
|
COSM1421475 COSM1421476 CA2742583 rs767684440 |
240 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA2742584 rs767684440 |
240 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA89854587 rs375653150 |
241 | P>R | No |
ClinGen ESP |
|
|
rs114869847 CA2742582 |
245 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1467805143 CA355843650 |
245 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA2742579 rs115485535 |
251 | I>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs373767629 CA2742580 |
251 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs753741084 CA2742578 |
253 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs763950262 CA2742577 COSM1484938 COSM1484937 |
256 | P>S | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 257 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2742576 rs559195466 |
257 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 257 | P>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 259 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1324052802 CA355843563 |
259 | P>R | No |
ClinGen gnomAD |
|
| TCGA novel | 260 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1171918339 CA355843535 |
263 | K>R | No |
ClinGen TOPMed |
|
|
CA2742572 rs182651924 |
266 | Y>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2742571 rs770565478 |
267 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA16040345 rs866449390 |
269 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs866449390 CA355843489 |
269 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA2742568 rs770229110 |
271 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA355843481 rs1250787586 |
271 | Y>H | No |
ClinGen gnomAD |
|
|
rs755391164 CA2742565 |
275 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs377183748 CA2742564 |
276 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373036001 CA2742561 |
277 | G>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1560047646 CA355843432 |
278 | M>I | No |
ClinGen Ensembl |
|
|
rs764055183 CA2742560 |
279 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs558126076 CA2742558 |
281 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2742559 rs558126076 |
281 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1489908260 CA355843412 |
282 | P>S | No |
ClinGen TOPMed |
|
|
CA89854583 rs981422304 |
283 | A>V | No |
ClinGen Ensembl |
|
|
CA355843400 rs1380872341 |
284 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs200659867 CA2742557 |
284 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs543445514 CA2742555 |
285 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2742554 rs144014237 CA355843377 |
287 | Q>H | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2742552 rs772562621 |
289 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA2742553 rs762479081 |
289 | P>S | No |
ClinGen ExAC |
|
|
CA2742551 rs376850706 |
290 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2742550 rs201447934 |
294 | Q>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA2742549 rs781711077 |
296 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs199529766 CA2742548 |
297 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs780199235 CA2742547 |
300 | C>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742546 rs780199235 |
300 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742545 rs201874364 |
301 | V>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs201874364 CA2742544 |
301 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2742542 rs757415664 |
302 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs772538831 CA2742528 |
307 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1274271382 CA355843237 |
307 | N>K | No |
ClinGen gnomAD |
|
|
rs1578542548 CA355843240 |
307 | N>T | No |
ClinGen Ensembl |
|
|
CA2742527 rs139046188 |
308 | C>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs779263596 CA2742526 |
309 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742525 rs757221699 |
310 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs749396927 CA2742524 |
310 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs1464091362 CA355843214 |
311 | S>F | No |
ClinGen TOPMed |
|
|
CA89854442 rs960183955 |
313 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1462723364 CA355843202 |
313 | M>V | No |
ClinGen gnomAD |
|
|
CA2742522 rs150747321 |
314 | R>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2742521 rs751594051 |
314 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA89854441 rs758276506 |
315 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758276506 CA2742519 |
315 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355843190 rs766071992 CA2742520 |
315 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC gnomAD NCI-TCGA |
|
CA2742518 rs749973761 |
316 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764956136 CA2742517 |
317 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA2742516 rs761262992 |
318 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1578542471 CA355843167 |
319 | S>P | No |
ClinGen Ensembl |
|
|
CA355843159 rs776148239 |
320 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776148239 CA2742515 |
320 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355843145 rs761150301 |
322 | H>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742513 rs761150301 |
322 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742514 rs763633676 |
322 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA355843136 rs1224872475 |
323 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs772320212 CA2742511 |
323 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs199656132 CA2742512 |
323 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746248776 CA2742510 |
324 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1052736998 CA89853712 |
326 | S>A | No |
ClinGen TOPMed |
|
|
CA2742493 rs768086594 |
327 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1302264839 CA355843104 |
327 | Y>H | No |
ClinGen gnomAD |
|
|
CA2742491 rs543262937 |
330 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1050122500 CA89853711 |
331 | P>R | No |
ClinGen TOPMed |
|
|
rs771465106 CA2742490 |
331 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA355843075 rs771465106 |
331 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA2742489 rs763251587 |
332 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM275013 COSM275012 rs769924564 CA2742487 |
337 | D>N | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1395826378 CA355843020 |
339 | C>Y | No |
ClinGen gnomAD |
|
| TCGA novel | 343 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355842987 rs1431724669 |
344 | R>K | No |
ClinGen gnomAD |
|
|
VAR_048951 rs2228269 CA2742470 |
348 | K>R | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs2228269 CA2742471 |
348 | K>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 349 | V>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355842939 rs1257362440 |
350 | K>E | No |
ClinGen gnomAD |
|
|
CA355842936 rs1345538245 |
350 | K>T | No |
ClinGen TOPMed |
|
|
rs1179613734 CA355842931 |
351 | Q>K | No |
ClinGen gnomAD |
|
|
rs770158979 CA2742469 |
353 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1203443297 CA355842885 |
357 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 361 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM209106 CA355842832 rs1274884541 |
365 | R>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
COSM1731299 rs1233658032 CA355842830 COSM1731298 |
365 | R>Q | liver [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1311407487 CA355842823 |
366 | G>V | No |
ClinGen TOPMed |
|
|
rs1291286379 CA355842822 |
367 | S>T | No |
ClinGen gnomAD |
|
|
rs75639224 CA89853699 |
373 | F>V | No |
ClinGen Ensembl |
|
|
rs1461706983 CA355842768 |
375 | V>F | No |
ClinGen gnomAD |
|
|
rs1461706983 CA355842767 |
375 | V>L | No |
ClinGen gnomAD |
|
|
CA2742465 rs745805257 |
376 | T>P | No |
ClinGen ExAC |
|
|
CA355842750 rs1398362632 |
378 | L>V | No |
ClinGen gnomAD |
|
|
rs1361584246 CA355842741 |
379 | D>G | No |
ClinGen gnomAD |
|
|
CA2742464 rs778889619 |
380 | D>A | No |
ClinGen ExAC gnomAD |
|
|
rs777358489 COSM1484934 COSM1484933 CA2742461 |
392 | R>Q | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA355842586 rs1250670377 |
401 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 404 | V>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1235270432 CA355842542 |
406 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA355842543 rs1256555691 |
406 | R>W | No |
ClinGen gnomAD |
|
|
CA355842533 rs1578535167 |
408 | W>G | No |
ClinGen Ensembl |
|
| TCGA novel | 411 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355842492 rs1578535163 |
413 | N>T | No |
ClinGen Ensembl |
|
|
CA355842483 rs1384213796 |
414 | R>Q | No |
ClinGen gnomAD |
|
|
CA355842414 rs1560044080 |
424 | R>C | No |
ClinGen Ensembl |
|
|
CA89853219 rs867599117 |
432 | K>E | No |
ClinGen Ensembl |
|
|
CA2742427 rs575192498 |
436 | Q>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA2742426 rs775581923 |
437 | K>N | No |
ClinGen ExAC |
|
|
CA2742407 rs767576093 |
444 | V>I | No |
ClinGen ExAC gnomAD |
|
|
COSM23333 CA89852898 rs267599722 |
445 | Y>C | skin [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1257894129 CA355842250 |
446 | K>R | No |
ClinGen gnomAD |
|
|
CA2742404 rs138236393 |
449 | C>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs761626996 CA2742403 |
452 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA2742401 rs768544405 |
457 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742402 rs768544405 |
457 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760388962 CA2742400 |
458 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA355842159 rs1225932263 |
459 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
COSM3427416 CA355842153 COSM3427417 rs1391685751 |
460 | P>L | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA355842158 rs1330279155 |
460 | P>T | No |
ClinGen gnomAD |
|
|
COSM1042266 COSM1042265 CA2742397 rs745384970 |
462 | N>S | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs962576898 CA89852895 |
463 | Q>H | No |
ClinGen TOPMed |
|
|
CA2742395 rs201212469 |
464 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs749844496 COSM1738861 COSM1738862 CA2742394 |
464 | R>H | Variant assessed as Somatic; 0.0 impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1178585908 CA355842089 |
470 | E>G | No |
ClinGen TOPMed |
|
| TCGA novel | 470 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1427101151 CA355842085 |
471 | S>T | No |
ClinGen TOPMed |
|
| TCGA novel | 472 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2742389 COSM1421463 rs138683352 COSM1421464 |
472 | E>K | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA355842069 rs1398948494 |
473 | C>Y | No |
ClinGen TOPMed |
|
|
rs766518204 CA355842057 |
474 | H>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 474 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs918949890 CA89852893 |
476 | S>N | No |
ClinGen Ensembl |
|
|
rs1002996879 CA89852892 |
477 | E>K | No |
ClinGen TOPMed |
|
|
CA355842026 rs1280846163 |
479 | D>A | No |
ClinGen gnomAD |
|
|
rs150223664 CA89852891 |
479 | D>N | No |
ClinGen ESP TOPMed |
|
|
rs184522009 CA2742384 |
480 | T>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2742385 rs753868848 |
480 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs1281944233 CA355842008 |
482 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1578533285 CA355842000 COSM1205751 COSM1205750 |
484 | T>A | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs771873806 CA2742381 |
486 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
rs908979960 CA89852889 |
489 | S>C | No |
ClinGen Ensembl |
|
|
rs1403599883 CA355841958 |
489 | S>R | No |
ClinGen gnomAD |
|
|
rs773704999 CA2742379 |
491 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA355841943 rs1578533262 |
492 | Y>S | No |
ClinGen Ensembl |
|
|
rs746030889 CA2742378 |
494 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2742377 rs749747970 |
495 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA2742376 rs773840485 |
496 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs770081575 CA2742375 |
498 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs550066362 CA2742374 COSM3392260 COSM3392259 |
498 | R>H | Variant assessed as Somatic; 0.0001386 impact. pancreas [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA355841898 rs1490469637 |
499 | C>Y | No |
ClinGen TOPMed |
|
|
rs138619028 CA2742372 |
502 | L>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs758570099 CA2742369 |
504 | Y>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA2742370 rs779983130 |
504 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs377733507 CA355841854 |
506 | E>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2742367 rs377733507 |
506 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA2742366 rs755924359 |
508 | F>V | No |
ClinGen ExAC gnomAD |
|
|
CA355841826 rs1331248488 |
510 | Y>D | No |
ClinGen TOPMed gnomAD |
No associated diseases with P41161
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| male germ-line stem cell asymmetric division | The self-renewing division of a germline stem cell in the male gonad, to produce a daughter stem cell and a daughter germ cell, which will divide to form the male gametes. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of glial cell proliferation | Any process that activates or increases the rate or extent of glial cell proliferation. |
| positive regulation of neuron differentiation | Any process that activates or increases the frequency, rate or extent of neuron differentiation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
40 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A1A4L6 | ETS2 | Protein C-ets-2 | Bos taurus (Bovine) | SS |
| Q2KIC2 | ETV1 | ETS translocation variant 1 | Bos taurus (Bovine) | SS |
| P15062 | ETS1 | Transforming protein p68/c-ets-1 | Gallus gallus (Chicken) | SS |
| P10157 | ETS2 | Protein C-ets-2 | Gallus gallus (Chicken) | SS |
| Q90837 | ERG | Transcriptional regulator Erg | Gallus gallus (Chicken) | SS |
| A2T762 | ETV3 | ETS translocation variant 3 | Pan troglodytes (Chimpanzee) | PR |
| Q04688 | Ets97D | DNA-binding protein Ets97D | Drosophila melanogaster (Fruit fly) | PR |
| Q9Y603 | ETV7 | Transcription factor ETV7 | Homo sapiens (Human) | SS |
| P41212 | ETV6 | Transcription factor ETV6 | Homo sapiens (Human) | SS |
| P78545 | ELF3 | ETS-related transcription factor Elf-3 | Homo sapiens (Human) | SS |
| Q9UKW6 | ELF5 | ETS-related transcription factor Elf-5 | Homo sapiens (Human) | EV |
| P32519 | ELF1 | ETS-related transcription factor Elf-1 | Homo sapiens (Human) | PR |
| Q99607 | ELF4 | ETS-related transcription factor Elf-4 | Homo sapiens (Human) | PR |
| Q06546 | GABPA | GA-binding protein alpha chain | Homo sapiens (Human) | SS |
| O95238 | SPDEF | SAM pointed domain-containing Ets transcription factor | Homo sapiens (Human) | PR |
| P50548 | ERF | ETS domain-containing transcription factor ERF | Homo sapiens (Human) | PR |
| P41162 | ETV3 | ETS translocation variant 3 | Homo sapiens (Human) | PR |
| P11308 | ERG | Transcriptional regulator ERG | Homo sapiens (Human) | EV |
| P43268 | ETV4 | ETS translocation variant 4 | Homo sapiens (Human) | EV |
| P50549 | ETV1 | ETS translocation variant 1 | Homo sapiens (Human) | EV |
| P41970 | ELK3 | ETS domain-containing protein Elk-3 | Homo sapiens (Human) | SS |
| P28324 | ELK4 | ETS domain-containing protein Elk-4 | Homo sapiens (Human) | EV |
| P19419 | ELK1 | ETS domain-containing protein Elk-1 | Homo sapiens (Human) | EV |
| P15036 | ETS2 | Protein C-ets-2 | Homo sapiens (Human) | EV |
| P14921 | ETS1 | Protein C-ets-1 | Homo sapiens (Human) | EV |
| P41971 | Elk3 | ETS domain-containing protein Elk-3 | Mus musculus (Mouse) | EV |
| Q00422 | Gabpa | GA-binding protein alpha chain | Mus musculus (Mouse) | EV |
| P41969 | Elk1 | ETS domain-containing protein Elk-1 | Mus musculus (Mouse) | PR |
| P28322 | Etv4 | ETS translocation variant 4 | Mus musculus (Mouse) | SS |
| P15037 | Ets2 | Protein C-ets-2 | Mus musculus (Mouse) | SS |
| P27577 | Ets1 | Protein C-ets-1 | Mus musculus (Mouse) | EV |
| P70459 | Erf | ETS domain-containing transcription factor ERF | Mus musculus (Mouse) | PR |
| P41158 | Elk4 | ETS domain-containing protein Elk-4 | Mus musculus (Mouse) | PR |
| P41164 | Etv1 | ETS translocation variant 1 | Mus musculus (Mouse) | SS |
| P81270 | Erg | Transcriptional regulator ERG | Mus musculus (Mouse) | SS |
| Q9CXC9 | Etv5 | ETS translocation variant 5 | Mus musculus (Mouse) | SS |
| P41156 | Ets1 | Protein C-ets-1 | Rattus norvegicus (Rat) | SS |
| A4GTP4 | Elk1 | ETS domain-containing protein Elk-1 | Rattus norvegicus (Rat) | PR |
| Q9PUQ1 | etv4 | ETS translocation variant 4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A3FEM2 | fev | Protein FEV | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDGFYDQQVP | FMVPGKSRSE | ECRGRPVIDR | KRKFLDTDLA | HDSEELFQDL | SQLQEAWLAE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AQVPDDEQFV | PDFQSDNLVL | HAPPPTKIKR | ELHSPSSELS | SCSHEQALGA | NYGEKCLYNY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CAYDRKPPSG | FKPLTPPTTP | LSPTHQNPLF | PPPQATLPTS | GHAPAAGPVQ | GVGPAPAPHS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LPEPGPQQQT | FAVPRPPHQP | LQMPKMMPEN | QYPSEQRFQR | QLSEPCHPFP | PQPGVPGDNR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PSYHRQMSEP | IVPAAPPPPQ | GFKQEYHDPL | YEHGVPGMPG | PPAHGFQSPM | GIKQEPRDYC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VDSEVPNCQS | SYMRGGYFSS | SHEGFSYEKD | PRLYFDDTCV | VPERLEGKVK | QEPTMYREGP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PYQRRGSLQL | WQFLVTLLDD | PANAHFIAWT | GRGMEFKLIE | PEEVARRWGI | QKNRPAMNYD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KLSRSLRYYY | EKGIMQKVAG | ERYVYKFVCD | PDALFSMAFP | DNQRPFLKAE | SECHLSEEDT |
| 490 | 500 | ||||
| LPLTHFEDSP | AYLLDMDRCS | SLPYAEGFAY |