P40450
Gene name |
BNR1 |
Protein name |
BNI1-related protein 1 |
Names |
|
Species |
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) |
KEGG Pathway |
sce:YIL159W |
EC number |
|
Protein Class |
|
Descriptions
(Annotation from UniProt)
The DAD domain regulates activation via by an autoinhibitory interaction with the GBD/FH3 domain. This autoinhibition is released upon competitive binding of an activated GTPase. The release of DAD allows the FH2 domain to then nucleate and elongate nonbranched actin filaments.
Autoinhibitory domains (AIDs)
Target domain |
94-490 (GBD/FH3 domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
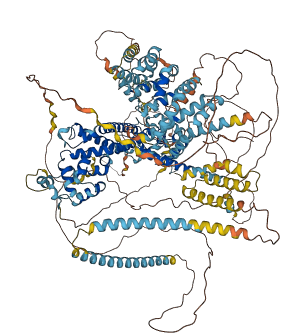
Autoinhibited structure

Activated structure

1 structures for P40450
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P40450-F1 | Predicted | AlphaFoldDB |
43 variants for P40450
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| s09-41898 | 25 | V>A | No | SGRP | |
| s09-42076 | 84 | N>K | No | SGRP | |
| s09-42275 | 151 | T>A | No | SGRP | |
| s09-42365 | 181 | T>A | No | SGRP | |
| s09-42785 | 321 | H>Y | No | SGRP | |
| s09-42866 | 348 | Q>E | No | SGRP | |
| s09-43004 | 394 | F>V | No | SGRP | |
| s09-43644 | 607 | S>N | No | SGRP | |
| s09-43664 | 614 | G>C | No | SGRP | |
| s09-43715 | 631 | A>T | No | SGRP | |
| s09-44120 | 766 | P>S | No | SGRP | |
| s09-44274 | 817 | A>V | No | SGRP | |
| s09-44331 | 836 | G>D | No | SGRP | |
| s09-44333 | 837 | P>S | No | SGRP | |
| s09-44342 | 840 | H>N | No | SGRP | |
| s09-44390 | 856 | S>P | No | SGRP | |
| s09-44414 | 864 | P>S | No | SGRP | |
| s09-44423 | 867 | T>A | No | SGRP | |
| s09-44424 | 867 | T>I | No | SGRP | |
| s09-44426 | 868 | D>N | No | SGRP | |
| s09-44433 | 870 | K>R | No | SGRP | |
| s09-44435 | 871 | P>A | No | SGRP | |
| s09-44438 | 872 | P>T | No | SGRP | |
| s09-44441 | 873 | P>S | No | SGRP | |
| s09-44444 | 874 | T>S | No | SGRP | |
| s09-44580 | 919 | I>T | No | SGRP | |
| s09-44586 | 921 | K>R | No | SGRP | |
| s09-44619 | 932 | R>K | No | SGRP | |
| s09-44634 | 937 | S>L | No | SGRP | |
| s09-44636 | 938 | I>V | No | SGRP | |
| s09-44643 | 940 | L>S | No | SGRP | |
| s09-44770 | 982 | M>I | No | SGRP | |
| s09-44944 | 1040 | L>F | No | SGRP | |
| s09-44979 | 1052 | A>V | No | SGRP | |
| s09-45053 | 1077 | I>V | No | SGRP | |
| s09-45096 | 1091 | K>R | No | SGRP | |
| s09-45203 | 1127 | I>V | No | SGRP | |
| s09-45243 | 1140 | V>G | No | SGRP | |
| s09-45363 | 1180 | I>T | No | SGRP | |
| s09-45559 | 1245 | K>N | No | SGRP | |
| s09-45691 | 1289 | D>E | No | SGRP | |
| s09-45705 | 1294 | S>N | No | SGRP | |
| s09-45746 | 1308 | K>E | No | SGRP |
No associated diseases with P40450
4 regional properties for P40450
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell division site | The eventual plane of cell division (also known as cell cleavage or cytokinesis) in a dividing cell. In Eukaryotes, the cleavage apparatus, composed of septin structures and the actomyosin contractile ring, forms along this plane, and the mitotic, or meiotic, spindle is aligned perpendicular to the division plane. In bacteria, the cell division site is generally located at mid-cell and is the site at which the cytoskeletal structure, the Z-ring, assembles. |
| cellular bud neck | The constriction between the mother cell and daughter cell (bud) in an organism that reproduces by budding. |
| mating projection tip | The apex of the mating projection in unicellular fungi exposed to mating pheromone; site of polarized growth. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| profilin binding | Binding to profilin, an actin-binding protein that forms a complex with G-actin and prevents it from polymerizing to form F-actin. |
| small GTPase binding | Binding to a small monomeric GTPase. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament bundle assembly | The assembly of actin filament bundles; actin filaments are on the same axis but may be oriented with the same or opposite polarities and may be packed with different levels of tightness. |
| actin nucleation | The initial step in the formation of an actin filament, in which actin monomers combine to form a new filament. Nucleation is slow relative to the subsequent addition of more monomers to extend the filament. |
| barbed-end actin filament capping | The binding of a protein or protein complex to the barbed (or plus) end of an actin filament, thus preventing the addition, exchange or removal of further actin subunits. |
| formin-nucleated actin cable assembly | The aggregation, arrangement and bonding together of a set of components to form a formin-nucleated actin cable. A formin-nucleated actin cable is an actin filament bundle that consists of short filaments organized into bundles of uniform polarity, and is nucleated by formins. |
| mitotic actomyosin contractile ring assembly | Any actomyosin contractile ring assembly that is involved in mitotic cytokinesis. |
| positive regulation of actin cytoskeleton reorganization | Any process that activates or increases the frequency, rate or extent of actin cytoskeleton reorganization. |
1 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P41832 | BNI1 | Protein BNI1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDSSPNKKTY | RYPRRSLSLH | ARDRVSEARK | LEELNLNDGL | VAAGLQLVGV | ALEKQGTGSH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IYMKQKNFSA | NDVSSSPMVS | EEVNGSEMDF | NPKCMPQDAS | LVERMFDELL | KDGTFFWGAA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YKNLQNISLR | RKWLLICKIR | SSNHWGKKKV | TSSTTYSTHL | ATNELAENAH | FLDGLVRNLS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TGGMKLSKAL | YKLEKFLRKQ | SFLQLFLKDE | IYLTTLIEKT | LPLISKELQF | VYLRCFKILM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NNPLARIRAL | HSEPLIRWFT | ELLTDQNSNL | KCQLLSMELL | LLLTYVEGST | GCELIWDQLS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ILFTDWLEWF | DKILADDIAI | HSSLYLNWNQ | LKIDYSTTFL | LLINSILQGF | NNKTALEILN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FLKKNNIHNT | ITFLELAYKD | DPNSVVIMEQ | IKQFKSKESA | IFDSMIKTTN | DTNSLHPTKD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| IARIESEPLC | LENCLLLKAK | DSPVEAPINE | IIQSLWKILD | SQKPYSESIK | LLKLINSLLF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YLIDSFQVST | NPSFDETLES | AENVDYVFQD | SVNKLLDSLQ | SDEIARRAVT | EIDDLNAKIS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| HLNEKLNLVE | NHDKDHLIAK | LDESESLISL | KTKEIENLKL | QLKATKKRLD | QITTHQRLYD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| QPPSLASSNL | SIAGSIIKNN | SHGNIIFQNL | AKKQQQQQKI | SLPKRSTSLL | KSKRVTSLSS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YLTDANNENE | SQNESEDKSK | DSLFQRSTST | INFNIPSMKN | ITNMQNVSLN | SILSELEFSN |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SLGTQPNYQS | SPVLSSVSSS | PKLFPRLSSD | SLDNGIQLVP | EVVKLPQLPP | PPPPPPPPPL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| PQSLLTEAEA | KPDGVSCIAA | PAPPPLPDLF | KTKTCGAVPP | PPPPPPLPES | LSMNKGPSNH |
| 850 | 860 | 870 | 880 | 890 | 900 |
| DLVTPPAPPL | PNGLLSSSSV | SINPTTTDLK | PPPTEKRLKQ | IHWDKVEDIK | DTLWEDTFQR |
| 910 | 920 | 930 | 940 | 950 | 960 |
| QETIKELQTD | GIFSQIEDIF | KMKSPTKIAN | KRNAESSIAL | SSNNGKSSNE | LKKISFLSRD |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LAQQFGINLH | MFSQLSDMEF | VMKVLNCDND | IVQNVNILKF | FCKEELVNIP | KSMLNKYEPY |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| SQGKDGKAVS | DLQRADRIFL | ELCINLRFYW | NARSKSLLTL | STYERDYYDL | IFKLQKIDDA |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| ISHLNRSPKF | KSLMFIITEI | GNHMNKRIVK | GIKLKSLTKL | AFVRSSIDQN | VSFLHFIEKV |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| IRIKYPDIYG | FVDDLKNIED | LGKISLEHVE | SECHEFHKKI | EDLVTQFQVG | KLSKEENLDP |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| RDQIIKKVKF | KINRAKTKSE | LLIGQCKLTL | IDLNKLMKYY | GEDPKDKESK | NEFFQPFIEF |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| LAMFKKCAKE | NIEKEEMERV | YEQRKSLLDM | RTSSNKKSNG | SDENDGEKVN | RDAVDLLISK |
| 1330 | 1340 | 1350 | 1360 | 1370 | |
| LREVKKDPEP | LRRRKSTKLN | EIAINVHEGD | VKTRKDEDHV | LLERTHAMLN | DIQNI |