P39073
Gene name |
SSN3 (ARE1, CDK8, GIG2, SRB10, UME5) |
Protein name |
Meiotic mRNA stability protein kinase SSN3 |
Names |
Cyclin-dependent kinase 8, Suppressor of RNA polymerase B SRB10 |
Species |
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) |
KEGG Pathway |
sce:YPL042C |
EC number |
2.7.11.22: Protein-serine/threonine kinases |
Protein Class |
|
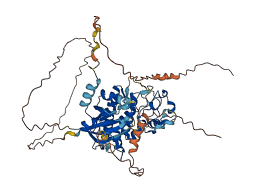
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
303-329 (Activation loop from InterPro)
Target domain |
75-463 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

3 structures for P39073
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 7KPV | EM | 380 A | A | 1-555 | PDB |
| 7KPX | EM | 440 A | A | 1-555 | PDB |
| AF-P39073-F1 | Predicted | AlphaFoldDB |
7 variants for P39073
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| s16-474585 | 40 | S>R | No | SGRP | |
| s16-474523 | 61 | R>K | No | SGRP | |
| s16-474367 | 113 | A>G | No | SGRP | |
| s16-474368 | 113 | A>T | No | SGRP | |
| s16-473816 | 297 | D>N | No | SGRP | |
| s16-473509 | 399 | I>T | No | SGRP | |
| s16-473467 | 413 | T>R | No | SGRP |
No associated diseases with P39073
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.22 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| CKM complex | Cyclin-dependent kinase complex which reversibly associates with the Mediator complex. In Saccharomyces cerevisiae it consists of SSN2, SSN3, SSN8 and SRB8. |
| mediator complex | A protein complex that interacts with the carboxy-terminal domain of the largest subunit of RNA polymerase II and plays an active role in transducing the signal from a transcription factor to the transcriptional machinery. The mediator complex is required for activation of transcription of most protein-coding genes, but can also act as a transcriptional corepressor. The Saccharomyces complex contains several identifiable subcomplexes: a head domain comprising Srb2, -4, and -5, Med6, -8, and -11, and Rox3 proteins; a middle domain comprising Med1, -4, and -7, Nut1 and -2, Cse2, Rgr1, Soh1, and Srb7 proteins; a tail consisting of Gal11p, Med2p, Pgd1p, and Sin4p; and a regulatory subcomplex comprising Ssn2, -3, and -8, and Srb8 proteins. Metazoan mediator complexes have similar modular structures and include homologs of yeast Srb and Med proteins. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| cyclin-dependent protein serine/threonine kinase activity | Cyclin-dependent catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| RNA polymerase II CTD heptapeptide repeat kinase activity | Catalysis of the reaction: ATP + RNA polymerase II large subunit CTD heptapeptide repeat (YSPTSPS) = ADP + H+ + phosphorylated RNA polymerase II. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| meiotic cell cycle | Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell via two nuclear divisions. |
| negative regulation of filamentous growth | Any process that decreases the frequency, rate or extent of the process in which a multicellular organism or a group of unicellular organisms grow in a threadlike, filamentous shape. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| nuclear-transcribed mRNA catabolic process, non-stop decay | The chemical reactions and pathways resulting in the breakdown of the transcript body of a nuclear-transcribed mRNA that is lacking a stop codon. |
| phosphorylation of RNA polymerase II C-terminal domain | The process of introducing a phosphate group on to an amino acid residue in the C-terminal domain of RNA polymerase II. Typically, this occurs during the transcription cycle and results in production of an RNA polymerase II enzyme where the carboxy-terminal domain (CTD) of the largest subunit is extensively phosphorylated, often referred to as hyperphosphorylated or the II(0) form. Specific types of phosphorylation within the CTD are usually associated with specific regions of genes, though there are exceptions. The phosphorylation state regulates the association of specific complexes such as the capping enzyme or 3'-RNA processing machinery to the elongating RNA polymerase complex. |
| positive regulation of transcription from RNA polymerase II promoter by galactose | Any process involving galactose that activates or increases the rate of transcription from an RNA polymerase II promoter. |
| protein destabilization | Any process that decreases the stability of a protein, making it more vulnerable to degradative processes or aggregation. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q03957 | CTK1 | CTD kinase subunit alpha | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q9VT57 | Cdk8 | Cyclin-dependent kinase 8 | Drosophila melanogaster (Fruit fly) | PR |
| Q9BWU1 | CDK19 | Cyclin-dependent kinase 19 | Homo sapiens (Human) | PR |
| P49336 | CDK8 | Cyclin-dependent kinase 8 | Homo sapiens (Human) | PR |
| Q8BWD8 | Cdk19 | Cyclin-dependent kinase 19 | Mus musculus (Mouse) | PR |
| Q8R3L8 | Cdk8 | Cyclin-dependent kinase 8 | Mus musculus (Mouse) | PR |
| Q6P3N6 | cdk8 | Cyclin-dependent kinase 8 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q8JH47 | cdk8 | Cyclin-dependent kinase 8 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MYNGKDRAQN | SYQPMYQRPM | QVQGQQQAQS | FVGKKNTIGS | VHGKAPMLMA | NNDVFTIGPY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RARKDRMRVS | VLEKYEVIGY | IAAGTYGKVY | KAKRQINSGT | NSANGSSLNG | TNAKIPQFDS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TQPKSSSSMD | MQANTNALRR | NLLKDEGVTP | GRIRTTREDV | SPHYNSQKQT | LIKKPLTVFY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AIKKFKTEKD | GVEQLHYTGI | SQSACREMAL | CRELHNKHLT | TLVEIFLERK | CVHMVYEYAE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HDLLQIIHFH | SHPEKRMIPP | RMVRSIMWQL | LDGVSYLHQN | WVLHRDLKPA | NIMVTIDGCV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KIGDLGLARK | FHNMLQTLYT | GDKVVVTIWY | RAPELLLGAR | HYTPAVDLWS | VGCIFAELIG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LQPIFKGEEA | KLDSKKTVPF | QVNQLQRILE | VLGTPDQKIW | PYLEKYPEYD | QITKFPKYRD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NLATWYHSAG | GRDKHALSLL | YHLLNYDPIK | RIDAFNALEH | KYFTESDIPV | SENVFEGLTY |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KYPARRIHTN | DNDIMNLGSR | TKNNTQASGI | TAGAAANALG | GLGVNRRILA | AAAAAAAAVS |
| 550 | |||||
| GNNASDEPSR | KKNRR |