P37231
Gene name |
PPARG (NR1C3) |
Protein name |
Peroxisome proliferator-activated receptor gamma |
Names |
PPAR-gamma, Nuclear receptor subfamily 1 group C member 3 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5468 |
EC number |
|
Protein Class |
|
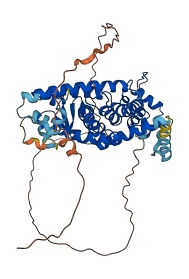
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

294 structures for P37231
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1FM6 | X-ray | 210 A | D/X | 234-505 | PDB |
| 1FM9 | X-ray | 210 A | D | 234-505 | PDB |
| 1I7I | X-ray | 235 A | A/B | 225-505 | PDB |
| 1K74 | X-ray | 230 A | D | 234-505 | PDB |
| 1KNU | X-ray | 250 A | A/B | 232-505 | PDB |
| 1NYX | X-ray | 265 A | A/B | 230-505 | PDB |
| 1PRG | X-ray | 220 A | A/B | 235-504 | PDB |
| 1RDT | X-ray | 240 A | D | 235-505 | PDB |
| 1WM0 | X-ray | 290 A | X | 232-505 | PDB |
| 1ZEO | X-ray | 250 A | A/B | 231-505 | PDB |
| 1ZGY | X-ray | 180 A | A | 234-505 | PDB |
| 2ATH | X-ray | 228 A | A/B | 235-505 | PDB |
| 2F4B | X-ray | 207 A | A/B | 235-505 | PDB |
| 2FVJ | X-ray | 199 A | A | 235-505 | PDB |
| 2G0G | X-ray | 254 A | A/B | 235-505 | PDB |
| 2G0H | X-ray | 230 A | A/B | 235-505 | PDB |
| 2GTK | X-ray | 210 A | A | 235-505 | PDB |
| 2HFP | X-ray | 200 A | A | 234-505 | PDB |
| 2HWQ | X-ray | 197 A | A/B | 235-505 | PDB |
| 2HWR | X-ray | 234 A | A/B | 235-505 | PDB |
| 2I4J | X-ray | 210 A | A/B | 223-504 | PDB |
| 2I4P | X-ray | 210 A | A/B | 223-504 | PDB |
| 2I4Z | X-ray | 225 A | A/B | 223-504 | PDB |
| 2OM9 | X-ray | 280 A | A/B/C/D | 232-505 | PDB |
| 2P4Y | X-ray | 225 A | A/B | 231-505 | PDB |
| 2POB | X-ray | 230 A | A/B | 234-505 | PDB |
| 2PRG | X-ray | 230 A | A/B | 235-505 | PDB |
| 2Q59 | X-ray | 220 A | A/B | 233-505 | PDB |
| 2Q5P | X-ray | 230 A | A/B | 233-505 | PDB |
| 2Q5S | X-ray | 205 A | A/B | 233-505 | PDB |
| 2Q61 | X-ray | 220 A | A/B | 233-505 | PDB |
| 2Q6R | X-ray | 241 A | A/B | 233-505 | PDB |
| 2Q6S | X-ray | 240 A | A/B | 233-505 | PDB |
| 2Q8S | X-ray | 230 A | A/B | 235-505 | PDB |
| 2QMV | NMR | - | A | 235-504 | PDB |
| 2VSR | X-ray | 205 A | A/B | 232-505 | PDB |
| 2VST | X-ray | 235 A | A/B | 232-505 | PDB |
| 2VV0 | X-ray | 255 A | A/B | 232-505 | PDB |
| 2VV1 | X-ray | 220 A | A/B | 232-505 | PDB |
| 2VV2 | X-ray | 275 A | A/B | 232-505 | PDB |
| 2VV3 | X-ray | 285 A | A/B | 232-505 | PDB |
| 2VV4 | X-ray | 235 A | A/B | 232-505 | PDB |
| 2XKW | X-ray | 202 A | A/B | 232-505 | PDB |
| 2YFE | X-ray | 200 A | A/B | 223-505 | PDB |
| 2ZK0 | X-ray | 236 A | A/B | 223-504 | PDB |
| 2ZK1 | X-ray | 261 A | A/B | 223-504 | PDB |
| 2ZK2 | X-ray | 226 A | A/B | 223-504 | PDB |
| 2ZK3 | X-ray | 258 A | A/B | 223-504 | PDB |
| 2ZK4 | X-ray | 257 A | A/B | 223-504 | PDB |
| 2ZK5 | X-ray | 245 A | A/B | 223-504 | PDB |
| 2ZK6 | X-ray | 241 A | A/B | 223-504 | PDB |
| 2ZNO | X-ray | 240 A | A/B | 223-504 | PDB |
| 2ZVT | X-ray | 190 A | A/B | 223-504 | PDB |
| 3ADS | X-ray | 225 A | A/B | 223-505 | PDB |
| 3ADT | X-ray | 270 A | A/B | 223-505 | PDB |
| 3ADU | X-ray | 277 A | A/B | 223-505 | PDB |
| 3ADV | X-ray | 227 A | A/B | 223-505 | PDB |
| 3ADW | X-ray | 207 A | A/B | 223-505 | PDB |
| 3ADX | X-ray | 195 A | A/B | 223-505 | PDB |
| 3AN3 | X-ray | 230 A | A/B | 223-504 | PDB |
| 3AN4 | X-ray | 230 A | A/B | 223-504 | PDB |
| 3B0Q | X-ray | 210 A | A/B | 231-504 | PDB |
| 3B0R | X-ray | 215 A | A/B | 231-504 | PDB |
| 3B1M | X-ray | 160 A | A | 234-505 | PDB |
| 3B3K | X-ray | 260 A | A/B | 223-504 | PDB |
| 3BC5 | X-ray | 227 A | A | 231-505 | PDB |
| 3CDP | X-ray | 280 A | A/B | 223-504 | PDB |
| 3CDS | X-ray | 265 A | A/B | 223-504 | PDB |
| 3CS8 | X-ray | 230 A | A | 234-504 | PDB |
| 3CWD | X-ray | 240 A | A/B | 236-505 | PDB |
| 3D6D | X-ray | 240 A | A/B | 223-504 | PDB |
| 3DZU | X-ray | 320 A | D | 102-505 | PDB |
| 3DZY | X-ray | 310 A | D | 102-505 | PDB |
| 3E00 | X-ray | 310 A | D | 102-505 | PDB |
| 3ET0 | X-ray | 240 A | A/B | 235-505 | PDB |
| 3ET3 | X-ray | 195 A | A | 235-505 | PDB |
| 3FEJ | X-ray | 201 A | A | 235-505 | PDB |
| 3FUR | X-ray | 230 A | A | 234-505 | PDB |
| 3G9E | X-ray | 230 A | A | 235-505 | PDB |
| 3GBK | X-ray | 230 A | A/B | 235-505 | PDB |
| 3H0A | X-ray | 210 A | D | 234-505 | PDB |
| 3HO0 | X-ray | 260 A | A/B | 223-504 | PDB |
| 3HOD | X-ray | 210 A | A/B | 223-504 | PDB |
| 3IA6 | X-ray | 231 A | A/B | 235-505 | PDB |
| 3K8S | X-ray | 255 A | A/B | 234-505 | PDB |
| 3KMG | X-ray | 210 A | A/D | 234-505 | PDB |
| 3LMP | X-ray | 190 A | A | 234-505 | PDB |
| 3NOA | X-ray | 198 A | A/B | 235-505 | PDB |
| 3OSI | X-ray | 270 A | A/B | 224-504 | PDB |
| 3OSW | X-ray | 255 A | A/B | 224-504 | PDB |
| 3PBA | X-ray | 230 A | A/B | 224-505 | PDB |
| 3PO9 | X-ray | 235 A | A/B | 224-505 | PDB |
| 3PRG | X-ray | 290 A | A | 232-505 | PDB |
| 3QT0 | X-ray | 250 A | A | 235-505 | PDB |
| 3R5N | X-ray | 200 A | A | 232-505 | PDB |
| 3R8A | X-ray | 241 A | A/B | 235-505 | PDB |
| 3R8I | X-ray | 230 A | A/B | 223-505 | PDB |
| 3S9S | X-ray | 255 A | A | 234-505 | PDB |
| 3SZ1 | X-ray | 230 A | A/B | 232-505 | PDB |
| 3T03 | X-ray | 210 A | A/B | 234-505 | PDB |
| 3TY0 | X-ray | 200 A | A/B | 231-505 | PDB |
| 3U9Q | X-ray | 152 A | A | 236-504 | PDB |
| 3V9T | X-ray | 165 A | A | 234-505 | PDB |
| 3V9V | X-ray | 160 A | A | 234-505 | PDB |
| 3V9Y | X-ray | 210 A | A | 234-505 | PDB |
| 3VJH | X-ray | 222 A | A/B | 223-504 | PDB |
| 3VJI | X-ray | 261 A | A/B | 223-504 | PDB |
| 3VN2 | X-ray | 218 A | A | 225-505 | PDB |
| 3VSO | X-ray | 200 A | A/B | 223-504 | PDB |
| 3VSP | X-ray | 240 A | A/B | 223-504 | PDB |
| 3WJ4 | X-ray | 195 A | A/B | 235-505 | PDB |
| 3WJ5 | X-ray | 189 A | A/B | 235-505 | PDB |
| 3WMH | X-ray | 210 A | A/B | 223-504 | PDB |
| 3X1H | X-ray | 230 A | A/B | 232-505 | PDB |
| 3X1I | X-ray | 240 A | A/B | 232-505 | PDB |
| 4A4V | X-ray | 200 A | A/B | 223-505 | PDB |
| 4A4W | X-ray | 200 A | A/B | 223-505 | PDB |
| 4CI5 | X-ray | 177 A | A/B | 234-505 | PDB |
| 4E4K | X-ray | 250 A | A/B | 223-505 | PDB |
| 4E4Q | X-ray | 250 A | A/B | 223-505 | PDB |
| 4EM9 | X-ray | 210 A | A/B | 235-505 | PDB |
| 4EMA | X-ray | 254 A | A/B | 235-505 | PDB |
| 4F9M | X-ray | 190 A | A | 234-505 | PDB |
| 4FGY | X-ray | 284 A | A | 235-504 | PDB |
| 4HEE | X-ray | 250 A | X | 235-505 | PDB |
| 4JAZ | X-ray | 285 A | A/B | 223-505 | PDB |
| 4JL4 | X-ray | 250 A | A/B | 223-505 | PDB |
| 4L96 | X-ray | 238 A | A | 235-505 | PDB |
| 4L98 | X-ray | 228 A | A/B | 235-505 | PDB |
| 4O8F | X-ray | 260 A | A/B | 223-505 | PDB |
| 4OJ4 | X-ray | 230 A | A | 232-505 | PDB |
| 4PRG | X-ray | 290 A | A/B/C/D | 235-504 | PDB |
| 4PVU | X-ray | 260 A | A/B | 223-505 | PDB |
| 4PWL | X-ray | 260 A | A/B | 223-505 | PDB |
| 4R06 | X-ray | 222 A | A/B | 233-505 | PDB |
| 4R2U | X-ray | 230 A | A/D | 231-505 | PDB |
| 4R6S | X-ray | 230 A | A/B | 231-505 | PDB |
| 4XLD | X-ray | 245 A | A | 231-505 | PDB |
| 4XTA | X-ray | 250 A | A/B | 232-505 | PDB |
| 4XUH | X-ray | 222 A | A/B | 232-505 | PDB |
| 4XUM | X-ray | 240 A | A/B | 232-505 | PDB |
| 4Y29 | X-ray | 198 A | A | 236-504 | PDB |
| 4YT1 | X-ray | 220 A | A/B | 223-504 | PDB |
| 5AZV | X-ray | 270 A | A/B | 232-505 | PDB |
| 5DSH | X-ray | 295 A | A | 223-505 | PDB |
| 5DV3 | X-ray | 275 A | A | 223-505 | PDB |
| 5DV6 | X-ray | 280 A | A | 223-505 | PDB |
| 5DV8 | X-ray | 275 A | A | 223-505 | PDB |
| 5DVC | X-ray | 230 A | A | 223-505 | PDB |
| 5DWL | X-ray | 220 A | A | 223-505 | PDB |
| 5F9B | X-ray | 225 A | A/B | 223-505 | PDB |
| 5GTN | X-ray | 185 A | A | 223-505 | PDB |
| 5GTO | X-ray | 210 A | A | 223-505 | PDB |
| 5GTP | X-ray | 235 A | A | 223-505 | PDB |
| 5HZC | X-ray | 200 A | A/B | 223-505 | PDB |
| 5JI0 | X-ray | 198 A | D | 234-505 | PDB |
| 5LSG | X-ray | 200 A | A/B | 223-505 | PDB |
| 5TTO | X-ray | 225 A | A/B | 233-505 | PDB |
| 5TWO | X-ray | 193 A | A | 234-505 | PDB |
| 5U5L | X-ray | 255 A | A/B | 233-505 | PDB |
| 5UGM | X-ray | 210 A | A/B | 235-505 | PDB |
| 5WQX | X-ray | 229 A | A/B | 232-505 | PDB |
| 5WR0 | X-ray | 285 A | A/B | 232-505 | PDB |
| 5WR1 | X-ray | 234 A | A/B | 232-505 | PDB |
| 5Y2O | X-ray | 180 A | A/B | 235-505 | PDB |
| 5Y2T | X-ray | 170 A | A/B | 235-505 | PDB |
| 5YCN | X-ray | 215 A | A | 223-505 | PDB |
| 5YCP | X-ray | 200 A | A | 223-505 | PDB |
| 5Z5S | X-ray | 180 A | A | 234-505 | PDB |
| 5Z6S | X-ray | 180 A | A | 234-505 | PDB |
| 6AD9 | X-ray | 220 A | A | 223-505 | PDB |
| 6AN1 | X-ray | 269 A | A/B | 224-505 | PDB |
| 6AUG | X-ray | 273 A | A/B | 231-505 | PDB |
| 6AVI | X-ray | 229 A | A/B | 231-505 | PDB |
| 6C1I | X-ray | 226 A | A/B | 231-504 | PDB |
| 6C5Q | X-ray | 240 A | A | 235-505 | PDB |
| 6C5T | X-ray | 275 A | A | 235-505 | PDB |
| 6D3E | X-ray | 240 A | A/B | 235-504 | PDB |
| 6D8X | X-ray | 190 A | A | 231-505 | PDB |
| 6D94 | X-ray | 190 A | A | 231-505 | PDB |
| 6DBH | X-ray | 260 A | A/B | 231-505 | PDB |
| 6DCU | X-ray | 295 A | A/B | 231-505 | PDB |
| 6DGL | X-ray | 195 A | A/B | 231-505 | PDB |
| 6DGO | X-ray | 310 A | A/B | 231-505 | PDB |
| 6DGP | X-ray | 310 A | A/B | 231-505 | PDB |
| 6DGQ | X-ray | 245 A | A/B | 231-505 | PDB |
| 6DGR | X-ray | 215 A | A/B | 231-505 | PDB |
| 6DH9 | X-ray | 270 A | A/B | 235-505 | PDB |
| 6DHA | X-ray | 188 A | A/B | 235-505 | PDB |
| 6E5A | X-ray | 240 A | A/B | 233-505 | PDB |
| 6ENQ | X-ray | 220 A | A/B | 224-505 | PDB |
| 6F2L | X-ray | 210 A | A/B | 223-505 | PDB |
| 6FZF | X-ray | 195 A | A/B | 231-505 | PDB |
| 6FZG | X-ray | 210 A | A | 231-505 | PDB |
| 6FZJ | X-ray | 201 A | A/B | 231-505 | PDB |
| 6FZP | X-ray | 230 A | A | 231-505 | PDB |
| 6FZY | X-ray | 310 A | A/B | 231-505 | PDB |
| 6ICJ | X-ray | 248 A | A | 234-505 | PDB |
| 6IJR | X-ray | 285 A | A/C | 223-505 | PDB |
| 6IJS | X-ray | 215 A | A | 232-505 | PDB |
| 6ILQ | X-ray | 241 A | A | 234-505 | PDB |
| 6IZM | X-ray | 180 A | A | 234-505 | PDB |
| 6IZN | X-ray | 175 A | A | 234-505 | PDB |
| 6JEY | X-ray | 220 A | A/B | 232-505 | PDB |
| 6JF0 | X-ray | 340 A | A/B | 232-505 | PDB |
| 6JQ7 | X-ray | 255 A | A | 223-505 | PDB |
| 6K0T | X-ray | 184 A | A/C | 223-505 | PDB |
| 6KTM | X-ray | 270 A | A | 223-505 | PDB |
| 6KTN | X-ray | 275 A | A | 223-505 | PDB |
| 6L89 | X-ray | 210 A | A/B | 223-505 | PDB |
| 6L8B | X-ray | 210 A | A/B | 223-505 | PDB |
| 6MCZ | X-ray | 210 A | A/B | 231-505 | PDB |
| 6MD0 | X-ray | 195 A | A/B | 231-505 | PDB |
| 6MD1 | X-ray | 220 A | A/B | 231-505 | PDB |
| 6MD2 | X-ray | 220 A | A/B | 231-505 | PDB |
| 6MD4 | X-ray | 224 A | A/B | 231-505 | PDB |
| 6MS7 | X-ray | 143 A | A | 234-505 | PDB |
| 6O67 | X-ray | 252 A | A/B | 231-505 | PDB |
| 6O68 | X-ray | 278 A | A/B | 231-505 | PDB |
| 6ONI | X-ray | 180 A | B | 231-505 | PDB |
| 6ONJ | X-ray | 230 A | A | 231-505 | PDB |
| 6PDZ | X-ray | 210 A | A/B | 231-505 | PDB |
| 6QJ5 | X-ray | 200 A | A/B | 223-505 | PDB |
| 6T1S | X-ray | 165 A | A | 231-505 | PDB |
| 6T1V | X-ray | 221 A | A | 231-505 | PDB |
| 6T6B | X-ray | 280 A | A | 231-505 | PDB |
| 6T9C | X-ray | 195 A | A/B | 223-505 | PDB |
| 6TDC | X-ray | 233 A | A | 235-505 | PDB |
| 6TSG | X-ray | 298 A | A | 231-505 | PDB |
| 6VZL | X-ray | 207 A | A/B | 231-505 | PDB |
| 6VZM | X-ray | 240 A | A/B | 231-505 | PDB |
| 6VZN | X-ray | 230 A | A/B | 231-505 | PDB |
| 6VZO | X-ray | 227 A | A/B | 231-505 | PDB |
| 6Y3U | X-ray | 262 A | A | 231-505 | PDB |
| 6ZLY | X-ray | 179 A | A/B | 223-505 | PDB |
| 7A7H | X-ray | 240 A | A | 231-505 | PDB |
| 7AHJ | X-ray | 210 A | A/B | 232-505 | PDB |
| 7AWC | X-ray | 174 A | A | 231-505 | PDB |
| 7AWD | X-ray | 193 A | A | 231-505 | PDB |
| 7CXE | X-ray | 250 A | A/B | 223-505 | PDB |
| 7CXF | X-ray | 235 A | A | 223-505 | PDB |
| 7CXG | X-ray | 188 A | A/B | 223-505 | PDB |
| 7CXH | X-ray | 230 A | A | 223-505 | PDB |
| 7CXI | X-ray | 230 A | A | 223-505 | PDB |
| 7CXJ | X-ray | 265 A | A | 223-505 | PDB |
| 7CXK | X-ray | 220 A | A | 223-505 | PDB |
| 7CXL | X-ray | 270 A | A | 223-505 | PDB |
| 7E0A | X-ray | 177 A | A | 230-505 | PDB |
| 7E2O | X-ray | 320 A | A/B | 232-505 | PDB |
| 7EFQ | X-ray | 230 A | A/B | 232-505 | PDB |
| 7JQG | X-ray | 215 A | A/B | 231-505 | PDB |
| 7LOT | X-ray | 229 A | A/B | 232-505 | PDB |
| 7P4E | X-ray | 240 A | A | 231-505 | PDB |
| 7QB1 | X-ray | 220 A | AAA/BBB | 223-505 | PDB |
| 7RLE | X-ray | 250 A | A/C | 231-505 | PDB |
| 7SQA | X-ray | 250 A | A/B | 231-505 | PDB |
| 7SQB | X-ray | 260 A | A | 231-505 | PDB |
| 7WGO | X-ray | 236 A | A | 231-505 | PDB |
| 7WGP | X-ray | 253 A | A | 231-505 | PDB |
| 7WGQ | X-ray | 243 A | A | 231-505 | PDB |
| 7WOX | X-ray | 320 A | A/B | 232-505 | PDB |
| 8ADF | X-ray | 213 A | A/B | 223-504 | PDB |
| 8AQM | X-ray | 230 A | A/B | 231-505 | PDB |
| 8AQN | X-ray | 190 A | A/B | 231-505 | PDB |
| 8ATY | X-ray | 190 A | A | 231-505 | PDB |
| 8ATZ | X-ray | 195 A | A | 231-505 | PDB |
| 8B8W | X-ray | 186 A | A/B | 231-505 | PDB |
| 8B8X | X-ray | 178 A | A/B | 231-505 | PDB |
| 8B8Y | X-ray | 200 A | A/B | 231-505 | PDB |
| 8B8Z | X-ray | 222 A | A/B | 231-505 | PDB |
| 8B90 | X-ray | 210 A | A/B | 231-505 | PDB |
| 8B91 | X-ray | 223 A | A/B | 231-505 | PDB |
| 8B92 | X-ray | 166 A | A/B | 231-505 | PDB |
| 8B93 | X-ray | 221 A | A/B | 231-505 | PDB |
| 8B94 | X-ray | 155 A | A/B | 231-505 | PDB |
| 8B95 | X-ray | 172 A | A/B | 231-505 | PDB |
| 8BF1 | X-ray | 136 A | A | 234-505 | PDB |
| 8BF2 | X-ray | 218 A | A/B | 234-505 | PDB |
| 8BFF | X-ray | 260 A | A/B/C | 234-505 | PDB |
| 8C0C | X-ray | 220 A | A/B | 223-504 | PDB |
| 8CPH | X-ray | 240 A | A/B | 231-505 | PDB |
| 8CPI | X-ray | 210 A | A | 231-505 | PDB |
| 8CPJ | X-ray | 240 A | A | 231-505 | PDB |
| 8DK4 | X-ray | 260 A | A | 234-505 | PDB |
| 8DKN | X-ray | 195 A | A | 234-505 | PDB |
| 8DKV | X-ray | 159 A | A | 234-505 | PDB |
| 8DSY | X-ray | 295 A | A/B | 234-505 | PDB |
| 8DSZ | X-ray | 250 A | A/B | 234-505 | PDB |
| 8HHP | X-ray | 245 A | A | 232-505 | PDB |
| 8HHQ | X-ray | 240 A | A/B | 232-505 | PDB |
| 8HUM | X-ray | 229 A | A | 231-505 | PDB |
| 8HUP | X-ray | 236 A | A | 231-505 | PDB |
| 8WFE | X-ray | 220 A | A/B | 234-505 | PDB |
| AF-P37231-F1 | Predicted | AlphaFoldDB |
337 variants for P37231
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000118044 rs1801282 VAR_010723 RCV000300998 RCV000336176 RCV000406618 RCV001516919 CA154763 |
12 | P>A | Obesity PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension significant independent determinant of CIMT; may protect from early atherosclerosis in subject at risk for diabetes; associated with BMI [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000008605 RCV000008604 CA119314 RCV000008606 VAR_016116 RCV000008607 rs1805192 |
40 | P>A | Diabetes mellitus, noninsulin-dependent, modifier of Body mass index, modifier of Intimal medial thickness of internal carotid artery, modifier of Obesity, modifier of [ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs2049559215 RCV001195982 |
47 | S>missing | Obesity [ClinVar] | Yes |
ClinVar dbSNP |
|
CA2258089 rs762280243 RCV001150848 RCV001147496 RCV001147497 |
49 | D>A | Obesity PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
RCV001150849 RCV001150850 rs777334819 COSM1484530 RCV001150851 RCV000501246 CA2258106 |
79 | E>K | Obesity Variant assessed as Somatic; 0.0 impact. PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension breast [ClinVar, NCI-TCGA, Cosmic] | Yes |
ClinGen cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV002512915 rs1800571 VAR_010724 RCV000008603 CA119312 |
113 | P>Q | Morbid obesity obesity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt TOPMed dbSNP gnomAD |
|
RCV000500158 CA351618202 rs1553643326 |
157 | E>G | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA351618328 RCV001146702 rs1211829538 RCV001759905 RCV001146703 RCV001147602 |
174 | D>G | Obesity PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000008608 rs587776687 |
186 | S>missing | Carcinoma of colon [ClinVar] | Yes |
ClinVar dbSNP |
|
rs121909245 CA119330 RCV000008620 |
190 | C>S | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs148195788 RCV000499572 CA70181654 |
194 | R>Q | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar ESP dbSNP |
|
rs1553647989 CA351618609 RCV000500715 |
212 | R>Q | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001262840 rs2050647141 |
235 | E>missing | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2050647473 RCV001253351 |
235 | E>missing | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001253577 rs150296212 |
240 | R>P | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV002559420 RCV001819854 rs150296212 RCV001148105 RCV001148104 RCV001148106 CA2258233 |
240 | R>Q | Obesity PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA351619329 RCV001174450 COSM1037601 rs1176746892 |
308 | R>C | Variant assessed as Somatic; 0.0 impact. Monogenic diabetes endometrium central_nervous_system [NCI-TCGA, ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar NCI-TCGA dbSNP gnomAD |
|
RCV001266805 rs2051075084 |
310 | F>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs121909242 RCV000008609 VAR_010725 CA250555 |
314 | Q>P | Carcinoma of colon colon cancer; sporadic; somatic mutation; loss of ligand-binding [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs28936407 VAR_010726 CA250559 RCV000008611 |
316 | R>H | Carcinoma of colon colon cancer; sporadic; somatic mutation; partial loss of ligand-binding [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt TOPMed dbSNP gnomAD |
|
CA119321 VAR_010727 rs72551362 RCV000008613 |
318 | V>M | PPARG-related familial partial lipodystrophy diabetes [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt ExAC dbSNP gnomAD |
|
rs1378972597 CA351619437 RCV000503151 |
324 | I>T | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV001174451 CA2258299 RCV000503975 RCV000918792 rs139894525 |
335 | V>L | Monogenic diabetes [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1553650477 RCV000499764 |
338 | D>missing | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000008610 CA250557 rs121909243 |
347 | K>* | Carcinoma of colon [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
VAR_022700 rs72551363 CA119326 RCV000008618 |
388 | F>L | PPARG-related familial partial lipodystrophy FPLD3 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000364024 RCV000301823 CA10614676 RCV000265474 rs886057902 |
402 | F>L | Obesity PPARG-related familial partial lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA119328 rs72551364 VAR_022701 |
425 | R>C | FPLD3 [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
RCV000501839 rs1553653993 CA351467669 |
451 | L>P | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000503538 rs770557781 |
454 | P>missing | PPARG-related familial partial lipodystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000664084 rs756523051 CA2258359 |
462 | K>R | Monogenic diabetes [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
VAR_010728 RCV000382908 CA119319 rs121909244 RCV000008612 RCV001851743 RCV001248978 RCV000288354 |
495 | P>L | Obesity PPARG-related familial partial lipodystrophy Lipodystrophy Diabetes Mellitus, Noninsulin-Dependent, with Acanthosis Nigricans and Hypertension diabetes [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
|
CA70177281 rs928058229 |
5 | L>V | No |
ClinGen Ensembl |
|
|
rs1447983492 CA351617021 |
7 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs768377660 CA2258022 |
8 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA351617039 rs1487931740 |
10 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs776891749 CA2258023 |
10 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351617043 rs765473307 |
11 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2258025 rs765473307 |
11 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763032994 CA2258026 |
14 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA70177282 rs1055170892 |
14 | S>N | No |
ClinGen Ensembl |
|
|
rs943725719 CA70177283 |
15 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs560683642 CA70177284 |
16 | S>F | No |
ClinGen gnomAD |
|
| TCGA novel | 16 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351617086 rs752068471 |
17 | F>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 20 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1273690252 CA351617107 |
20 | T>R | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 21 | L>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs572991054 CA351617109 |
21 | L>V | No |
ClinGen 1000Genomes ExAC TOPMed |
|
|
CA351617134 rs1320029787 |
25 | I>V | No |
ClinGen gnomAD |
|
|
rs753139831 CA351617140 |
26 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753139831 CA2258031 |
26 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1575040061 CA351617152 |
27 | Q>H | No |
ClinGen Ensembl |
|
|
rs780983036 CA2258076 |
30 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA351617354 rs1335787704 |
31 | M>I | No |
ClinGen gnomAD |
|
|
rs1289387362 CA351617347 |
31 | M>V | No |
ClinGen gnomAD |
|
|
rs1010142067 CA70180279 |
32 | V>F | No |
ClinGen TOPMed |
|
|
CA351617356 rs1010142067 |
32 | V>I | No |
ClinGen TOPMed |
|
|
CA351617360 rs1215582219 |
33 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA351617378 rs1186361559 |
35 | E>G | No |
ClinGen TOPMed |
|
|
CA351617375 rs1246297562 |
35 | E>Q | No |
ClinGen gnomAD |
|
|
rs1464788240 CA351617384 |
36 | M>L | No |
ClinGen gnomAD |
|
|
CA2258077 rs748119466 |
36 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA2258079 rs777492588 |
39 | W>R | No |
ClinGen ExAC gnomAD |
|
|
rs749089217 CA2258080 |
41 | T>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 41 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351617418 rs749089217 |
41 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA70180280 rs375411329 |
42 | N>S | No |
ClinGen Ensembl |
|
|
rs375308989 CA2258082 |
43 | F>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2258083 rs143370807 |
45 | I>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2258084 rs772234905 |
47 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772234905 CA351617460 |
47 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs141797536 CA2258086 |
48 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs141797536 CA2258087 |
48 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs138038967 CA2258090 |
49 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2258088 rs777067241 |
49 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA351617472 rs1235373823 |
50 | L>H | No |
ClinGen gnomAD |
|
|
rs1235373823 CA351617474 |
50 | L>R | No |
ClinGen gnomAD |
|
|
rs751292113 CA2258091 |
50 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA2258092 rs372494308 |
51 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs149324518 CA2258094 |
52 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA70180282 rs907515750 |
53 | M>L | No |
ClinGen Ensembl |
|
|
CA351617505 rs756002554 |
55 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA2258095 rs756002554 |
55 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs1441242852 CA351617510 |
56 | H>D | No |
ClinGen gnomAD |
|
|
COSM1495287 CA351617515 rs1187225480 |
56 | H>Q | kidney [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA351617513 rs1559515493 |
56 | H>R | No |
ClinGen Ensembl |
|
|
CA70180284 rs868377089 |
57 | S>F | No |
ClinGen Ensembl |
|
|
CA70180283 rs868377089 |
57 | S>Y | No |
ClinGen Ensembl |
|
| TCGA novel | 59 | S>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA70180285 rs369178039 |
62 | I>N | No |
ClinGen ESP |
|
|
CA70180286 rs369178039 |
62 | I>T | No |
ClinGen ESP |
|
|
CA2258096 rs139400454 |
62 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs757147631 CA2258099 |
66 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA2258100 rs778975987 |
67 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1429812309 CA351617588 |
68 | V>F | No |
ClinGen TOPMed |
|
|
rs1413870551 CA351617602 |
70 | F>V | No |
ClinGen gnomAD |
|
|
CA70180287 rs145995117 |
71 | S>C | No |
ClinGen ESP |
|
|
rs1391398513 CA351617614 |
72 | S>G | No |
ClinGen TOPMed |
|
|
CA70180289 rs201978569 |
75 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2258103 rs201978569 |
75 | T>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs201978569 CA351617634 |
75 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA351617648 rs1559515590 |
77 | H>R | No |
ClinGen Ensembl |
|
|
CA351617656 rs1286678629 |
78 | Y>C | No |
ClinGen gnomAD |
|
| TCGA novel | 80 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351617696 rs375156001 |
84 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2258108 rs375156001 |
84 | T>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1207461391 CA351617701 |
85 | R>G | No |
ClinGen TOPMed |
|
|
rs1196196054 CA351617703 |
85 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs773848402 CA2258109 |
87 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs759176486 CA2258110 |
90 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs140915035 CA2258111 |
91 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2258113 rs760238759 |
92 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA2258112 rs369959243 |
92 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1183297709 CA351617754 |
93 | Y>F | No |
ClinGen gnomAD |
|
|
CA2258114 rs764018224 |
94 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA70180290 rs892566497 |
94 | K>R | No |
ClinGen Ensembl |
|
|
rs1477623791 CA351617766 |
95 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs181976291 CA70180291 |
96 | D>N | No |
ClinGen 1000Genomes gnomAD |
|
|
CA351617779 rs1427549233 |
97 | L>V | No |
ClinGen gnomAD |
|
|
rs756980631 CA2258116 |
98 | K>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 101 | E>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2258118 rs750218440 |
102 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA351617822 rs1329435104 |
103 | Q>R | No |
ClinGen gnomAD |
|
|
CA2258119 rs758581958 |
104 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA351617850 rs1234040540 |
105 | A>E | No |
ClinGen TOPMed |
|
|
CA70180439 rs984243899 |
105 | A>T | No |
ClinGen gnomAD |
|
|
CA351617853 rs1262418036 |
106 | I>V | No |
ClinGen gnomAD |
|
|
rs765091444 CA2258138 |
108 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1165659288 CA351617903 |
114 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs751871408 CA2258142 |
116 | Y>D | No |
ClinGen ExAC gnomAD |
|
|
CA2258143 rs755081984 |
117 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs148037954 CA351617927 |
117 | S>F | No |
ClinGen TOPMed |
|
|
rs148037954 CA70180441 |
117 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1363321156 CA351617947 |
120 | T>I | No |
ClinGen gnomAD |
|
|
CA351617951 rs1296488686 |
121 | Q>E | No |
ClinGen TOPMed |
|
|
rs781368576 CA2258144 |
121 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1436657901 CA351617959 |
122 | L>F | No |
ClinGen TOPMed |
|
|
rs1028136694 CA351617970 |
123 | Y>* | No |
ClinGen TOPMed gnomAD |
|
|
CA2258145 rs748205580 |
124 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs756575717 CA2258146 |
124 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1214500245 CA351618020 |
130 | P>L | No |
ClinGen gnomAD |
|
|
rs1452375580 CA351618015 |
130 | P>T | No |
ClinGen TOPMed |
|
|
rs1237800473 CA351618029 |
132 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
CA351618031 rs1261231626 |
132 | N>S | No |
ClinGen gnomAD |
|
|
rs1261231626 CA351618030 |
132 | N>T | No |
ClinGen gnomAD |
|
|
CA70180444 rs267599576 |
133 | S>F | No |
ClinGen ExAC |
|
|
rs267599576 CA2258150 |
133 | S>Y | No |
ClinGen ExAC |
|
|
CA351618053 rs1193598966 |
135 | M>I | No |
ClinGen TOPMed |
|
|
rs1474976112 CA351618057 |
136 | A>E | No |
ClinGen gnomAD |
|
|
rs1256582642 CA351618054 |
136 | A>T | No |
ClinGen gnomAD |
|
|
rs549579646 CA2258153 |
137 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA2258155 rs765206131 |
140 | R>C | No |
ClinGen ExAC |
|
|
CA2258156 rs773089349 |
140 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2258157 rs762777341 |
141 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1259224734 CA351618103 |
143 | G>E | No |
ClinGen TOPMed |
|
|
CA351618113 rs1455807261 |
144 | D>E | No |
ClinGen gnomAD |
|
|
rs1346246354 CA351618106 |
144 | D>N | No |
ClinGen gnomAD |
|
|
CA70180445 rs796683680 |
151 | Y>* | No |
ClinGen Ensembl |
|
|
CA351618160 rs1354592503 |
151 | Y>C | No |
ClinGen gnomAD |
|
|
rs1575087860 CA351618171 |
153 | V>F | No |
ClinGen Ensembl |
|
|
CA2258161 rs767853271 |
158 | G>E | No |
ClinGen ExAC TOPMed |
|
|
RCV000503040 rs1553645235 CA351618245 |
161 | G>V | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA2258191 rs747804355 |
162 | F>V | No |
ClinGen ExAC gnomAD |
|
|
CA351618262 rs1170672782 |
164 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA2258193 rs777103332 |
165 | R>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 167 | I>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2258195 rs149064546 |
167 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2258194 rs748808980 |
167 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA351618303 rs1214981971 |
170 | K>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1222604363 CA351618297 |
170 | K>Q | No |
ClinGen TOPMed |
|
|
rs1315048063 CA351618315 |
172 | I>S | No |
ClinGen TOPMed |
|
|
rs771815282 CA2258198 |
172 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775382056 CA2258199 |
173 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA2258200 rs781526093 |
175 | R>K | No |
ClinGen ExAC gnomAD |
|
|
COSM1037500 rs764381653 CA2258201 |
178 | L>I | Variant assessed as Somatic; 0.0006007 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA2258202 rs753857970 |
181 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs149802972 CA70181652 |
183 | H>P | No |
ClinGen Ensembl |
|
|
rs1475289630 CA351618388 |
183 | H>Y | No |
ClinGen gnomAD |
|
|
rs1575106051 CA351618394 |
184 | K>E | No |
ClinGen Ensembl |
|
|
CA351618409 COSM1642070 rs1421126930 |
186 | S>G | stomach [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
| rs587776687 | 186 | S>V | Variant assessed as Somatic; 4.624e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs971658739 CA70181653 |
189 | K>N | No |
ClinGen Ensembl |
|
|
CA119332 rs121909246 |
194 | R>W | No |
ClinGen gnomAD |
|
|
CA351618517 rs1462603839 |
201 | V>M | No |
ClinGen gnomAD |
|
| TCGA novel | 204 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1325850074 CA351618548 |
205 | H>R | No |
ClinGen gnomAD |
|
|
CA2258223 rs765346374 |
208 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs766913119 CA351618642 |
217 | E>* | No |
ClinGen ExAC gnomAD |
|
|
rs766913119 CA2258226 |
217 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA351618673 rs1346411548 |
221 | L>V | No |
ClinGen gnomAD |
|
|
CA351618683 rs1233945910 |
222 | L>F | No |
ClinGen gnomAD |
|
| TCGA novel | 223 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs573789959 COSM2152381 CA70183036 |
223 | A>V | Variant assessed as Somatic; 0.0 impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
|
rs755059777 CA70183037 |
227 | S>G | No |
ClinGen Ensembl |
|
|
CA351618712 rs1193699370 |
227 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1421791772 CA351618728 |
229 | I>N | No |
ClinGen TOPMed |
|
|
rs763647203 CA2258228 |
230 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351618732 rs763647203 |
230 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763647203 CA2258229 |
230 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756994321 CA2258230 |
233 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351618755 rs756994321 |
233 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2258231 rs778527431 |
234 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA351618773 rs1453730891 |
236 | S>A | No |
ClinGen gnomAD |
|
|
CA70183039 rs569100208 |
237 | A>T | No |
ClinGen gnomAD |
|
|
rs1273579652 CA351618792 |
239 | L>F | No |
ClinGen TOPMed |
|
|
CA2258232 rs745376706 |
240 | R>W | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 243 | A>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs780100410 CA2258234 |
246 | L>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA70183040 rs956182399 |
247 | Y>C | No |
ClinGen Ensembl |
|
|
CA2258235 rs746778926 |
248 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768538430 CA2258236 |
249 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA351618858 rs1369335310 |
250 | Y>H | No |
ClinGen gnomAD |
|
|
rs1559527858 CA351618868 |
251 | I>T | No |
ClinGen Ensembl |
|
|
rs776809755 CA2258237 |
251 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1010744856 CA70183041 |
255 | P>L | No |
ClinGen Ensembl |
|
|
rs903976924 CA70183042 |
259 | A>V | No |
ClinGen TOPMed |
|
|
CA2258241 rs763106561 |
263 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1339677544 CA351618949 |
264 | I>V | No |
ClinGen gnomAD |
|
|
CA70183043 rs76480583 |
266 | T>K | No |
ClinGen Ensembl |
|
| TCGA novel | 267 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 268 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2258243 rs774987355 |
268 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs971821381 CA70183044 |
270 | T>A | No |
ClinGen Ensembl |
|
|
CA2258246 rs767945316 |
271 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1194161227 CA351618995 |
271 | D>G | No |
ClinGen gnomAD |
|
|
rs756904247 CA351619002 |
272 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756904247 CA2258247 |
272 | K>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA70184211 rs200116144 |
274 | P>L | No |
ClinGen 1000Genomes gnomAD |
|
|
rs4135352 CA351619095 |
275 | F>L | No |
ClinGen gnomAD |
|
|
rs147996578 CA2258279 |
276 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 277 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351619111 rs1311453246 |
278 | Y>C | No |
ClinGen gnomAD |
|
|
CA70184213 rs979542536 |
278 | Y>H | No |
ClinGen gnomAD |
|
|
rs935360461 CA70184215 |
281 | N>S | No |
ClinGen Ensembl |
|
| TCGA novel | 284 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs745949478 CA2258280 |
286 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1370501806 CA351619210 |
291 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs776209503 CA2258282 |
292 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA351619248 rs1355789926 |
296 | T>N | No |
ClinGen gnomAD |
|
|
CA351619244 rs1575141139 |
296 | T>P | No |
ClinGen Ensembl |
|
|
rs1347367417 CA351619260 |
298 | L>R | No |
ClinGen gnomAD |
|
|
CA351619270 rs1324719941 |
300 | E>Q | No |
ClinGen TOPMed |
|
|
CA351619279 rs1239470707 |
301 | Q>K | No |
ClinGen gnomAD |
|
|
rs765948672 CA2258287 |
302 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA351619304 rs1575141215 |
304 | E>G | No |
ClinGen Ensembl |
|
|
CA351619316 rs1575141221 |
306 | A>P | No |
ClinGen Ensembl |
|
|
CA2258288 rs751019400 |
308 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA70184217 rs751019400 |
308 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA70184218 rs111400984 |
309 | I>T | No |
ClinGen Ensembl |
|
|
rs767437202 CA2258290 |
312 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1364613522 CA351619383 |
316 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA351619421 rs1440763451 |
322 | Q>R | No |
ClinGen gnomAD |
|
|
rs1438652871 CA351619439 |
324 | I>M | No |
ClinGen gnomAD |
|
|
rs757444052 CA2258294 |
326 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs758594107 CA351619483 |
331 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758594107 CA2258297 |
331 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747713716 CA2258298 |
335 | V>Y | No |
ClinGen ExAC |
|
|
rs938504972 CA70184220 |
338 | D>G | No |
ClinGen TOPMed |
|
|
CA351619543 rs1274014409 |
340 | N>D | No |
ClinGen TOPMed |
|
|
CA351619584 rs1253000910 |
346 | L>F | No |
ClinGen gnomAD |
|
|
CA70184222 rs375109042 |
350 | V>G | No |
ClinGen ESP gnomAD |
|
|
rs929756683 CA70184221 |
350 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
COSM1037602 RCV000501649 rs530007199 CA2258306 |
352 | E>K | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ClinVar ExAC NCI-TCGA dbSNP gnomAD |
|
rs530007199 CA70184223 |
352 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA351619651 rs145566594 |
356 | T>A | No |
ClinGen gnomAD |
|
|
rs145566594 CA70184224 |
356 | T>P | No |
ClinGen gnomAD |
|
|
rs1325132092 CA351619661 |
357 | M>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs767061053 CA2258307 |
357 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1381954870 CA351619670 |
359 | A>P | No |
ClinGen TOPMed |
|
|
CA351619677 rs1438276181 |
360 | S>A | No |
ClinGen gnomAD |
|
|
rs780294601 CA2258308 |
361 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2258309 rs377025335 |
363 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371491950 CA2258311 |
369 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA351619748 rs1215517121 |
370 | S>F | No |
ClinGen gnomAD |
|
|
CA2258313 rs141683496 |
371 | E>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1273044250 CA351619756 |
372 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA2258314 rs750726364 |
375 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs758720345 CA2258315 |
378 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs888116131 CA70184225 |
379 | E>G | No |
ClinGen TOPMed |
|
|
CA351619804 RCV000503417 rs1553650533 |
379 | E>K | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1443416779 CA351619820 |
381 | L>V | No |
ClinGen TOPMed |
|
|
CA351619830 rs1559533293 |
382 | K>N | No |
ClinGen Ensembl |
|
|
CA351619838 rs1240742379 CA351619839 |
383 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
rs780238349 CA351619846 |
385 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA2258317 rs140204299 |
385 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
rs188622140 CA70184227 |
388 | F>C | No |
ClinGen Ensembl |
|
|
CA351619869 rs1182239192 |
389 | G>S | No |
ClinGen TOPMed |
|
|
rs1233791403 CA351619911 |
394 | P>H | No |
ClinGen TOPMed |
|
|
CA351619977 rs1407041548 |
403 | N>S | No |
ClinGen gnomAD |
|
|
CA2258322 rs773968047 |
404 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs745473692 CA2258323 |
404 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351619991 rs1400424764 |
406 | E>Q | No |
ClinGen gnomAD |
|
|
CA70184229 rs968233147 |
408 | D>V | No |
ClinGen Ensembl |
|
|
rs771718768 CA2258324 |
410 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs148844673 CA2258325 |
410 | S>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1231461827 CA351620033 |
411 | D>E | No |
ClinGen gnomAD |
|
| TCGA novel | 416 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351620082 rs1283709828 |
419 | I>V | No |
ClinGen gnomAD |
|
|
CA2258344 rs771700358 |
426 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1160189065 CA351467362 |
427 | G>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 429 | L>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351467407 rs1393014897 |
431 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA351467426 rs1179543971 |
432 | K>M | No |
ClinGen TOPMed |
|
|
RCV000500036 CA351467428 rs1437186245 |
432 | K>N | No |
ClinGen ClinVar TOPMed dbSNP |
|
|
CA2258345 rs779748988 |
433 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA69571948 rs896261519 |
434 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs746500248 CA2258346 |
434 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 436 | D>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA69571957 rs1011908622 |
437 | I>M | No |
ClinGen TOPMed |
|
|
rs768103795 CA2258347 |
437 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 439 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2258351 rs367723779 |
444 | A>V | No |
ClinGen ESP ExAC TOPMed |
|
|
CA351467604 rs1442054619 |
446 | E>K | No |
ClinGen TOPMed |
|
|
RCV000118042 rs587780424 CA231410 COSM1669972 |
450 | K>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
CA69572022 rs962987706 |
451 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA2258356 rs759638890 |
454 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs186728157 CA2258357 |
455 | E>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs186728157 CA69572062 |
455 | E>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA351467736 rs1457864825 |
457 | S>L | No |
ClinGen gnomAD |
|
|
rs1251138457 CA351467743 |
458 | Q>H | No |
ClinGen gnomAD |
|
|
CA351467771 rs1156628228 |
463 | L>M | No |
ClinGen TOPMed |
|
|
rs951599607 CA69572079 |
467 | M>I | No |
ClinGen Ensembl |
|
| TCGA novel | 472 | Q>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351467967 rs1479145908 |
475 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs62243567 CA69572109 |
477 | H>N | No |
ClinGen Ensembl |
|
|
CA351468011 rs1333944746 |
478 | V>G | No |
ClinGen gnomAD |
|
|
CA351468004 rs1466241270 |
478 | V>M | No |
ClinGen gnomAD |
|
|
rs975376703 CA69572121 |
485 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs746597317 CA2258363 |
486 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA351468112 rs1302451905 |
487 | T>A | No |
ClinGen gnomAD |
|
|
CA2258364 COSM1642075 rs201781800 |
487 | T>M | stomach [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC gnomAD |
|
rs747703645 CA2258366 |
488 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA69572133 rs914519822 |
492 | S>R | No |
ClinGen gnomAD |
|
|
CA351468206 rs1200047582 |
494 | H>P | No |
ClinGen gnomAD |
|
|
CA351468217 rs121909244 |
495 | P>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 499 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
4 associated diseases with P37231
[MIM: 601665]: Obesity (OBESITY)
A condition characterized by an increase of body weight beyond the limitation of skeletal and physical requirements, as the result of excessive accumulation of body fat. {ECO:0000269|PubMed:9753710}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry.
[MIM: 604367]: Lipodystrophy, familial partial, 3 (FPLD3)
A form of lipodystrophy characterized by marked loss of subcutaneous fat from the extremities. Facial adipose tissue may be increased, decreased or normal. Affected individuals show an increased preponderance of insulin resistance, diabetes mellitus and dyslipidemia. {ECO:0000269|PubMed:11788685, ECO:0000269|PubMed:12453919}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 137800]: Glioma 1 (GLM1)
Gliomas are benign or malignant central nervous system neoplasms derived from glial cells. They comprise astrocytomas and glioblastoma multiforme that are derived from astrocytes, oligodendrogliomas derived from oligodendrocytes and ependymomas derived from ependymocytes. {ECO:0000269|PubMed:10851250}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry. Polymorphic PPARG alleles have been found to be significantly over-represented among a cohort of American patients with sporadic glioblastoma multiforme suggesting a possible contribution to disease susceptibility.
Without disease ID
- A condition characterized by an increase of body weight beyond the limitation of skeletal and physical requirements, as the result of excessive accumulation of body fat. {ECO:0000269|PubMed:9753710}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry.
- A form of lipodystrophy characterized by marked loss of subcutaneous fat from the extremities. Facial adipose tissue may be increased, decreased or normal. Affected individuals show an increased preponderance of insulin resistance, diabetes mellitus and dyslipidemia. {ECO:0000269|PubMed:11788685, ECO:0000269|PubMed:12453919}. Note=The disease is caused by variants affecting the gene represented in this entry.
- Gliomas are benign or malignant central nervous system neoplasms derived from glial cells. They comprise astrocytomas and glioblastoma multiforme that are derived from astrocytes, oligodendrogliomas derived from oligodendrocytes and ependymomas derived from ependymocytes. {ECO:0000269|PubMed:10851250}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry. Polymorphic PPARG alleles have been found to be significantly over-represented among a cohort of American patients with sporadic glioblastoma multiforme suggesting a possible contribution to disease susceptibility.
Functions
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| RNA polymerase II transcription regulator complex | A transcription factor complex that acts at a regulatory region of a gene transcribed by RNA polymerase II. |
28 GO annotations of molecular function
| Name | Definition |
|---|---|
| alpha-actinin binding | Binding to alpha-actinin, one of a family of proteins that cross-link F-actin as antiparallel homodimers. Alpha-actinin has a molecular mass of 93-103 KDa; at the N-terminus there are two calponin homology domains, at the C-terminus there are two EF-hands. These two domains are connected by the rod domain. This domain is formed by triple-helical spectrin repeats. |
| arachidonic acid binding | Binding to arachidonic acid, a straight chain fatty acid with 20 carbon atoms and four double bonds per molecule. Arachidonic acid is the all-Z-(5,8,11,14)-isomer. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA binding domain binding | Binding to a protein's DNA binding domain (DBD). |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| double-stranded DNA binding | Binding to double-stranded DNA. |
| E-box binding | Binding to an E-box, a DNA motif with the consensus sequence CANNTG that is found in the promoters of a wide array of genes expressed in neurons, muscle and other tissues. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| identical protein binding | Binding to an identical protein or proteins. |
| LBD domain binding | Binding to a protein's ligand binding domain (LBD) domain, found in nuclear receptors. In general, the LBDs consist of three layers comprised of twelve alpha-helices and several beta-strands that are organized around a lipophilic ligand-binding pocket. |
| nuclear receptor activity | A DNA-binding transcription factor activity regulated by binding to a ligand that modulates the transcription of specific gene sets transcribed by RNA polymerase II. Nuclear receptor ligands are usually lipid-based (such as a steroid hormone) and the binding of the ligand to its receptor often occurs in the cytoplasm, which leads to its tranlocation to the nucleus. |
| nuclear retinoid X receptor binding | Binding to a nuclear retinoid X receptor. |
| nucleic acid binding | Binding to a nucleic acid. |
| peptide binding | Binding to a peptide, an organic compound comprising two or more amino acids linked by peptide bonds. |
| prostaglandin receptor activity | Combining with a prostaglandin (PG) to initiate a change in cell activity. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein self-association | Binding to a domain within the same polypeptide. |
| R-SMAD binding | Binding to a receptor-regulated SMAD signaling protein. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| STAT family protein binding | Binding to a member of the signal transducers and activators of transcription (STAT) protein family. STATs are, as the name indicates, both signal transducers and transcription factors. STATs are activated by cytokines and some growth factors and thus control important biological processes including cell growth, cell differentiation, apoptosis and immune responses. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
| transcription coregulator binding | Binding to a transcription coregulator, a protein involved in regulation of transcription via protein-protein interactions with transcription factors and other transcription regulatory proteins. Cofactors do not bind DNA directly, but rather mediate protein-protein interactions between regulatory transcription factors and the basal transcription machinery. |
| zinc ion binding | Binding to a zinc ion (Zn). |
77 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that initiates the activity of the inactive enzyme cysteine-type endopeptidase in the context of an apoptotic process. |
| BMP signaling pathway | The series of molecular signals initiated by the binding of a member of the BMP (bone morphogenetic protein) family to a receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell fate commitment | The commitment of cells to specific cell fates and their capacity to differentiate into particular kinds of cells. Positional information is established through protein signals that emanate from a localized source within a cell (the initial one-cell zygote) or within a developmental field. |
| cell maturation | A developmental process, independent of morphogenetic (shape) change, that is required for a cell to attain its fully functional state. |
| cellular response to hypoxia | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cellular response to low-density lipoprotein particle stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a low-density lipoprotein particle stimulus. |
| epithelial cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of an epithelial cell, any of the cells making up an epithelium. |
| fatty acid metabolic process | The chemical reactions and pathways involving fatty acids, aliphatic monocarboxylic acids liberated from naturally occurring fats and oils by hydrolysis. |
| glucose homeostasis | Any process involved in the maintenance of an internal steady state of glucose within an organism or cell. |
| hormone-mediated signaling pathway | The series of molecular signals mediated by the detection of a hormone. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| lipid homeostasis | Any process involved in the maintenance of an internal steady state of lipid within an organism or cell. |
| lipid metabolic process | The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. |
| lipoprotein transport | The directed movement of any conjugated, water-soluble protein in which the nonprotein group consists of a lipid or lipids, into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| long-chain fatty acid transport | The directed movement of long-chain fatty acids into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. A long-chain fatty acid is a fatty acid with a chain length between C13 and C22. |
| macrophage derived foam cell differentiation | The process in which a monocyte acquires the specialized features of a foam cell. A foam cell is a type of cell containing lipids in small vacuoles and typically seen in atherosclerotic lesions, as well as other conditions. |
| monocyte differentiation | The process in which a relatively unspecialized myeloid precursor cell acquires the specialized features of a monocyte. |
| mRNA transcription by RNA polymerase II | The cellular synthesis of messenger RNA (mRNA) from a DNA template by RNA polymerase II, originating at an RNA polymerase II promoter. |
| negative regulation of angiogenesis | Any process that stops, prevents, or reduces the frequency, rate or extent of angiogenesis. |
| negative regulation of blood vessel endothelial cell migration | Any process that stops, prevents, or reduces the frequency, rate or extent of the migration of the endothelial cells of blood vessels. |
| negative regulation of BMP signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the BMP signaling pathway. |
| negative regulation of cardiac muscle hypertrophy in response to stress | Any process that stops, prevents or reduces the frequency, rate or extent of cardiac muscle hypertrophy in response to stress. |
| negative regulation of cellular response to transforming growth factor beta stimulus | Any process that stops, prevents or reduces the frequency, rate or extent of cellular response to transforming growth factor beta stimulus. |
| negative regulation of cholesterol storage | Any process that decreases the rate or extent of cholesterol storage. Cholesterol storage is the accumulation and maintenance in cells or tissues of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. |
| negative regulation of connective tissue replacement involved in inflammatory response wound healing | Any process that stops, prevents or reduces the frequency, rate or extent of connective tissue replacement involved in inflammatory response wound healing. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of inflammatory response | Any process that stops, prevents, or reduces the frequency, rate or extent of the inflammatory response. |
| negative regulation of interferon-gamma-mediated signaling pathway | Any process that decreases the rate, frequency or extent of an interferon-gamma-mediated signaling pathway. |
| negative regulation of lipid storage | Any process that decreases the rate, frequency or extent of lipid storage. Lipid storage is the accumulation and maintenance in cells or tissues of lipids, compounds soluble in organic solvents but insoluble or sparingly soluble in aqueous solvents. Lipid reserves can be accumulated during early developmental stages for mobilization and utilization at later stages of development. |
| negative regulation of macrophage derived foam cell differentiation | Any process that decreases the rate, frequency or extent of macrophage derived foam cell differentiation. Macrophage derived foam cell differentiation is the process in which a macrophage acquires the specialized features of a foam cell. A foam cell is a type of cell containing lipids in small vacuoles and typically seen in atherosclerotic lesions, as well as other conditions. |
| negative regulation of MAP kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of MAP kinase activity. |
| negative regulation of miRNA transcription | Any process that stops, prevents or reduces the frequency, rate or extent of microRNA (miRNA) gene transcription. |
| negative regulation of miRNA-mediated gene silencing | A process that decreases the rate, frequency, or extent of gene silencing by a microRNA (miRNA). |
| negative regulation of mitochondrial fission | Any process that decreases the rate, frequency or extent of mitochondrial fission. Mitochondrial fission is the division of a mitochondrion within a cell to form two or more separate mitochondrial compartments. |
| negative regulation of osteoblast differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of osteoblast differentiation. |
| negative regulation of pathway-restricted SMAD protein phosphorylation | Any process that decreases the rate, frequency or extent of pathway-restricted SMAD protein phosphorylation. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
| negative regulation of receptor signaling pathway via STAT | Any process that stops, prevents or reduces the frequency, rate or extent of receptor signaling via STAT. |
| negative regulation of sequestering of triglyceride | Any process that decreases the rate, frequency or extent of sequestering of triglyceride. Triglyceride sequestration is the process of binding or confining any triester of glycerol such that it is separated from other components of a biological system. |
| negative regulation of signaling receptor activity | Any process that stops, prevents or reduces the frequency, rate or extent of a signaling receptor activity. |
| negative regulation of SMAD protein signal transduction | Any process that decreases the rate, frequency or extent of the SMAD protein signaling pathway. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
| negative regulation of smooth muscle cell proliferation | Any process that stops, prevents or reduces the rate or extent of smooth muscle cell proliferation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transforming growth factor beta receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of any TGF-beta receptor signaling pathway. |
| negative regulation of vascular associated smooth muscle cell proliferation | Any process that stops, prevents or reduces the frequency, rate or extent of vascular smooth muscle cell proliferation. |
| negative regulation of vascular endothelial cell proliferation | Any process that stops, prevents or reduces the frequency, rate or extent of vascular endothelial cell proliferation. |
| peroxisome proliferator activated receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to any of the peroxisome proliferator activated receptors (alpha, beta or gamma) in the nuclear membrane, and ending with the initiation or termination of the transcription of target genes. |
| placenta development | The process whose specific outcome is the progression of the placenta over time, from its formation to the mature structure. The placenta is an organ of metabolic interchange between fetus and mother, partly of embryonic origin and partly of maternal origin. |
| positive regulation of adiponectin secretion | Any process that activates or increases the frequency, rate or extent of the regulated release of adiponectin from a cell. |
| positive regulation of adipose tissue development | Any process that activates or increases the frequency, rate or extent of adipose tissue development. |
| positive regulation of cholesterol efflux | Any process that increases the frequency, rate or extent of cholesterol efflux. Cholesterol efflux is the directed movement of cholesterol, cholest-5-en-3-beta-ol, out of a cell or organelle. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of DNA-binding transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of fat cell differentiation | Any process that activates or increases the frequency, rate or extent of adipocyte differentiation. |
| positive regulation of fatty acid metabolic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving fatty acids. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of low-density lipoprotein receptor activity | Any process that activates or increases the frequency, rate or extent of low-density lipoprotein receptor activity. |
| positive regulation of miRNA transcription | Any process that activates or increases the frequency, rate or extent of microRNA (miRNA) gene transcription. |
| positive regulation of pathway-restricted SMAD protein phosphorylation | Any process that increases the rate, frequency or extent of pathway-restricted SMAD protein phosphorylation. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
| positive regulation of SMAD protein signal transduction | Any process that increases the rate, frequency or extent of SMAD protein signal transduction. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of vascular associated smooth muscle cell apoptotic process | Any process that activates or increases the frequency, rate or extent of vascular associated smooth muscle cell apoptotic process. |
| regulation of blood pressure | Any process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure. |
| regulation of cellular response to insulin stimulus | Any process that modulates the frequency, rate or extent of cellular response to insulin stimulus. |
| regulation of cholesterol transporter activity | Any process that modulates the rate, frequency, or extent of cholesterol transporter activity. |
| regulation of circadian rhythm | Any process that modulates the frequency, rate or extent of a circadian rhythm. A circadian rhythm is a biological process in an organism that recurs with a regularity of approximately 24 hours. |
| regulation of lipid metabolic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving lipids. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| response to lipid | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipid stimulus. |
| response to nutrient | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus. |
| retinoic acid receptor signaling pathway | The series of molecular signals generated as a consequence of a retinoic acid receptor binding to one of its physiological ligands. |
| rhythmic process | Any process pertinent to the generation and maintenance of rhythms in the physiology of an organism. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| white fat cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a white adipocyte, an animal connective tissue cell involved in energy storage. White adipocytes have cytoplasmic lipids arranged in a unique vacuole. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9TTR7 | NR2F2 | COUP transcription factor 2 | Bos taurus (Bovine) | PR |
| O18971 | PPARG | Peroxisome proliferator-activated receptor gamma | Bos taurus (Bovine) | PR |
| Q90733 | NR2F2 | COUP transcription factor 2 | Gallus gallus (Chicken) | PR |
| P68306 | THRB | Thyroid hormone receptor beta | Gallus gallus (Chicken) | PR |
| A7X8B3 | PGR | Progesterone receptor | Pan troglodytes (Chimpanzee) | SS |
| P10588 | NR2F6 | Nuclear receptor subfamily 2 group F member 6 | Homo sapiens (Human) | PR |
| P06401 | PGR | Progesterone receptor | Homo sapiens (Human) | EV |
| P10589 | NR2F1 | COUP transcription factor 1 | Homo sapiens (Human) | PR |
| P24468 | NR2F2 | COUP transcription factor 2 | Homo sapiens (Human) | PR |
| P49116 | NR2C2 | Nuclear receptor subfamily 2 group C member 2 | Homo sapiens (Human) | PR |
| O75469 | NR1I2 | Nuclear receptor subfamily 1 group I member 2 | Homo sapiens (Human) | PR |
| Q14995 | NR1D2 | Nuclear receptor subfamily 1 group D member 2 | Homo sapiens (Human) | PR |
| P35396 | Ppard | Peroxisome proliferator-activated receptor delta | Mus musculus (Mouse) | PR |
| P43136 | Nr2f6 | Nuclear receptor subfamily 2 group F member 6 | Mus musculus (Mouse) | PR |
| P43135 | Nr2f2 | COUP transcription factor 2 | Mus musculus (Mouse) | PR |
| Q00175 | Pgr | Progesterone receptor | Mus musculus (Mouse) | SS |
| Q9Z0Y9 | Nr1h3 | Oxysterols receptor LXR-alpha | Mus musculus (Mouse) | PR |
| O62807 | PPARG | Peroxisome proliferator-activated receptor gamma | Sus scrofa (Pig) | PR |
| O09018 | Nr2f2 | COUP transcription factor 2 | Rattus norvegicus (Rat) | PR |
| Q63449 | Pgr | Progesterone receptor | Rattus norvegicus (Rat) | SS |
| Q8SQ01 | NR1I2 | Nuclear receptor subfamily 1 group I member 2 | Macaca mulatta (Rhesus macaque) | PR |
| G5EFF5 | daf-12 | Nuclear hormone receptor family member daf-12 | Caenorhabditis elegans | PR |
| O45460 | nhr-54 | Nuclear hormone receptor family member nhr-54 | Caenorhabditis elegans | PR |
| Q21006 | nhr-34 | Nuclear hormone receptor family member nhr-34 | Caenorhabditis elegans | PR |
| O17928 | nhr-52 | Nuclear hormone receptor family member nhr-52 | Caenorhabditis elegans | PR |
| Q21878 | nhr-1 | Nuclear hormone receptor family member nhr-1 | Caenorhabditis elegans | PR |
| O18141 | nhr-79 | Nuclear hormone receptor family member nhr-79 | Caenorhabditis elegans | PR |
| Q20765 | nhr-7 | Nuclear hormone receptor family member nhr-7 | Caenorhabditis elegans | PR |
| Q6PH18 | nr2f1b | Nuclear receptor subfamily 2 group F member 1-B | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGETLGDSPI | DPESDSFTDT | LSANISQEMT | MVDTEMPFWP | TNFGISSVDL | SVMEDHSHSF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DIKPFTTVDF | SSISTPHYED | IPFTRTDPVV | ADYKYDLKLQ | EYQSAIKVEP | ASPPYYSEKT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QLYNKPHEEP | SNSLMAIECR | VCGDKASGFH | YGVHACEGCK | GFFRRTIRLK | LIYDRCDLNC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RIHKKSRNKC | QYCRFQKCLA | VGMSHNAIRF | GRMPQAEKEK | LLAEISSDID | QLNPESADLR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ALAKHLYDSY | IKSFPLTKAK | ARAILTGKTT | DKSPFVIYDM | NSLMMGEDKI | KFKHITPLQE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QSKEVAIRIF | QGCQFRSVEA | VQEITEYAKS | IPGFVNLDLN | DQVTLLKYGV | HEIIYTMLAS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LMNKDGVLIS | EGQGFMTREF | LKSLRKPFGD | FMEPKFEFAV | KFNALELDDS | DLAIFIAVII |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LSGDRPGLLN | VKPIEDIQDN | LLQALELQLK | LNHPESSQLF | AKLLQKMTDL | RQIVTEHVQL |
| 490 | 500 | ||||
| LQVIKKTETD | MSLHPLLQEI | YKDLY |