P36015
Gene name |
YKT6 (YKL196C) |
Protein name |
Synaptobrevin homolog YKT6 |
Names |
|
Species |
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) |
KEGG Pathway |
sce:YKL196C |
EC number |
|
Protein Class |
SYNAPTOBREVIN HOMOLOG YKT6 (PTHR45806) |
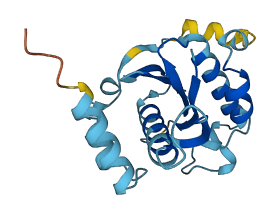
Descriptions
Most SNAREs are permanently anchored to membranes, but the dually lipidated SNARE Ykt6 is found both on intracellular membranes and in the cytoplasm. The SNARE core is autoinhibited by the N-terminal longin domain.
The dynamics of Ykt6 are believed to be governed by the reversible palmitoylation of the protein, which cycles Ykt6 between intracellular membranes and the cytoplasm. Palmitoylation of Ykt6 increases the partition coefficient of Ykt6 to membranes, thereby shifting some populations of the protein from the cytosol to cellular membranes. Palmitoylation-mediated membrane insertion will further shift the conformational equilibrium of Ykt6 to the open state. The specific membrane localization and the amount of membrane-associated Ykt6 are predicted to be affected by its reversible palmitoylation machinery in cells, as the majority of Ykt6 exists in the unpalmitoylated cytosolic form
Autoinhibitory domains (AIDs)
Target domain |
140-200 (v-SNARE coiled-coil homology) |
Relief mechanism |
PTM |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure
Activated structure

4 structures for P36015
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1H8M | NMR | - | A | 1-140 | PDB |
| 1IOU | NMR | - | A | 1-140 | PDB |
| 3BW6 | X-ray | 250 A | A | 1-140 | PDB |
| AF-P36015-F1 | Predicted | AlphaFoldDB |
No variants for P36015
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P36015 | |||||
No associated diseases with P36015
No regional properties for P36015
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P36015 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR45806 | SYNAPTOBREVIN HOMOLOG YKT6 |
| PANTHER Subfamily | PTHR45806:SF1 | SYNAPTOBREVIN HOMOLOG YKT6 |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| fungal-type vacuole | A vacuole that has both lytic and storage functions. The fungal vacuole is a large, membrane-bounded organelle that functions as a reservoir for the storage of small molecules (including polyphosphate, amino acids, several divalent cations (e.g. calcium), other ions, and other small molecules) as well as being the primary compartment for degradation. It is an acidic compartment, containing an ensemble of acid hydrolases. At least in S. cerevisiae, there are indications that the morphology of the vacuole is variable and correlated with the cell cycle, with logarithmically growing cells having a multilobed, reticulated vacuole, while stationary phase cells contain a single large structure. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| SNARE complex | A protein complex involved in membrane fusion; a stable ternary complex consisting of a four-helix bundle, usually formed from one R-SNARE and three Q-SNAREs with an ionic layer sandwiched between hydrophobic layers. One well-characterized example is the neuronal SNARE complex formed of synaptobrevin 2, syntaxin 1a, and SNAP-25. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| palmitoyltransferase activity | Catalysis of the transfer of a palmitoyl (CH3-14-CO-) group to an acceptor molecule. |
| SNAP receptor activity | Acting as a marker to identify a membrane and interacting selectively with one or more SNAREs on another membrane to mediate membrane fusion. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| amphisome-lysosome fusion | The process in which amphisomes fuse with a vacuole (yeast) or lysosome (e.g. mammals and insects). In the case of yeast, inner membrane-bounded structures (autophagic bodies) appear in the vacuole. Fusion provides an acidic environment and digestive function to the interior of the amphisome. |
| endoplasmic reticulum to Golgi vesicle-mediated transport | The directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi. |
| Golgi to endosome transport | The directed movement of substances from the Golgi to early sorting endosomes. Clathrin vesicles transport substances from the trans-Golgi to endosomes. |
| Golgi vesicle fusion to target membrane | The joining of the lipid bilayer membrane around a Golgi transport vesicle to the target lipid bilayer membrane. |
| intra-Golgi vesicle-mediated transport | The directed movement of substances within the Golgi, mediated by small transport vesicles. These either fuse with the cis-Golgi or with each other to form the membrane stacks known as the cis-Golgi reticulum (network). |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| vacuole fusion, non-autophagic | The fusion of two vacuole membranes to form a single vacuole. |
| vesicle fusion | Fusion of the membrane of a transport vesicle with its target membrane. |
| vesicle fusion with Golgi apparatus | The joining of the lipid bilayer membrane around a vesicle to the lipid bilayer membrane around the Golgi. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3T000 | YKT6 | Synaptobrevin homolog YKT6 | Bos taurus (Bovine) | SS |
| O15498 | YKT6 | Synaptobrevin homolog YKT6 | Homo sapiens (Human) | SS |
| Q9CQW1 | Ykt6 | Synaptobrevin homolog YKT6 | Mus musculus (Mouse) | SS |
| Q5EGY4 | Ykt6 | Synaptobrevin homolog YKT6 | Rattus norvegicus (Rat) | EV |
| Q9LVM9 | YKT62 | VAMP-like protein YKT62 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZRD6 | YKT61 | VAMP-like protein YKT61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q6P816 | ykt6 | Synaptobrevin homolog YKT6 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7ZUN8 | ykt6 | Synaptobrevin homolog YKT6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRIYYIGVFR | SGGEKALELS | EVKDLSQFGF | FERSSVGQFM | TFFAETVASR | TGAGQRQSIE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EGNYIGHVYA | RSEGICGVLI | TDKEYPVRPA | YTLLNKILDE | YLVAHPKEEW | ADVTETNDAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KMKQLDTYIS | KYQDPSQADA | IMKVQQELDE | TKIVLHKTIE | NVLQRGEKLD | NLVDKSESLT |
| 190 | |||||
| ASSKMFYKQA | KKSNSCCIIM |