P35716
Gene name |
SOX11 |
Protein name |
Transcription factor SOX-11 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6664 |
EC number |
|
Protein Class |
SOX TRANSCRIPTION FACTOR (PTHR10270) |
Descriptions
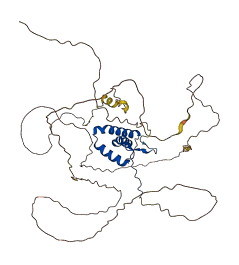
Sox11 acts as a transcriptional activator of TEAD2 by binding to its gene promoter. This protein possesses intrinsically disordered regions (IDRs) with large negative charge, some of which involve a consecutive sequence of aspartate (D) or glutamate (E) residues, known as D/E repeats. These D/E repeats can cause autoinhibition through intramolecular electrostatic interaction with HMG boxes and modulate binding to DNA. This autoinhibited state can transition into the uninhibited complex with DNA through an electrostatically driven induced-fit process, which accelerates the target DNA search kinetics in the presence of non-functional high-affinity ligands ('decoys').
Autoinhibitory domains (AIDs)
Target domain |
38-129 (HMG box domain) |
Relief mechanism |
|
Assay |
Peptide inhibitor test |
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

5 structures for P35716
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6T78 | X-ray | 250 A | A/B | 33-138 | PDB |
| 6T7A | EM | 370 A | K | 33-138 | PDB |
| 6T7C | EM | 400 A | K/L | 33-138 | PDB |
| 6T7D | EM | 440 A | K | 33-138 | PDB |
| AF-P35716-F1 | Predicted | AlphaFoldDB |
423 variants for P35716
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| VAR_088026 | 29 | C>del | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088027 | 47 | G>S | IDDMOH; uncertain significance [UniProt] | Yes | UniProt |
| VAR_088028 | 48 | H>D | IDDMOH; uncertain significance [UniProt] | Yes | UniProt |
| VAR_088029 | 49 | I>N | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088030 | 50 | K>N | IDDMOH; decreased function in positive regulation of DNA-templated transcription; compared to the wild-type, the mutant induces a severely reduced GDF5 promoter activation using an in vitro reporter system [UniProt] | Yes | UniProt |
| VAR_088031 | 50 | K>Q | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088032 | 51 | R>G | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088033 | 51 | R>L | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088034 | 51 | R>Q | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088035 | 51 | R>W | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088036 | 52 | P>L | IDDMOH [UniProt] | Yes | UniProt |
|
TCGA novel VAR_088037 |
53 | M>I | Variant assessed as Somatic; MODERATE impact. IDDMOH [NCI-TCGA, UniProt] | Yes |
NCI-TCGA UniProt |
| VAR_088038 | 53 | M>R | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088039 | 53 | M>V | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088040 | 55 | A>T | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088041 | 56 | F>L | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088042 | 57 | M>T | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088043 | 59 | W>del | IDDMOH [UniProt] | Yes | UniProt |
|
CA163236 VAR_071461 RCV000128429 rs587777480 |
60 | S>P | Intellectual disability, autosomal dominant 27 IDDMOH; decreases transcriptional activity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
VAR_088044 COSM1021939 |
64 | R>C | Variant assessed as Somatic; MODERATE impact. IDDMOH [NCI-TCGA, UniProt] | Yes |
NCI-TCGA Cosmic UniProt |
| VAR_088045 | 64 | R>L | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088046 | 64 | R>P | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088047 | 64 | R>S | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088048 | 75 | H>D | IDDMOH [UniProt] | Yes | UniProt |
|
RCV001246232 VAR_088049 rs1665660543 RCV003150825 |
76 | N>D | Intellectual disability, autosomal dominant 27 IDDMOH [ClinVar, UniProt] | Yes |
ClinVar dbSNP UniProt |
|
rs1553327809 RCV000624664 CA345866265 |
80 | S>C | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
TCGA novel VAR_088050 |
84 | G>S | Variant assessed as Somatic; MODERATE impact. IDDMOH [NCI-TCGA, UniProt] | Yes |
NCI-TCGA UniProt |
| VAR_088051 | 87 | W>R | IDDMOH [UniProt] | Yes | UniProt |
|
rs1558373252 RCV000735210 |
98 | F>missing | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_088052 | 98 | F>L | IDDMOH; uncertain significance [UniProt] | Yes | UniProt |
|
rs1064794628 VAR_088053 CA16617744 RCV003150820 RCV000481795 |
100 | R>P | Intellectual disability, autosomal dominant 27 IDDMOH [ClinVar, UniProt] | Yes |
ClinGen ClinVar dbSNP gnomAD UniProt |
|
rs1665661372 RCV001089956 VAR_088054 COSM4094946 |
102 | A>V | Intellectual disability, autosomal dominant 27 Variant assessed as Somatic; MODERATE impact. IDDMOH [ClinVar, NCI-TCGA, UniProt] | Yes |
NCI-TCGA Cosmic ClinVar dbSNP UniProt |
| VAR_088055 | 106 | R>P | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088056 | 109 | H>P | IDDMOH [UniProt] | Yes | UniProt |
| VAR_088057 | 113 | Y>H | IDDMOH; decreased function in positive regulation of DNA-templated transcription; compared to the wild-type, the mutant induces reduced GDF5 promoter activation using an in vitro reporter system; no effect on nuclear localization [UniProt] | Yes | UniProt |
|
VAR_071462 rs587777479 RCV000128428 CA163235 |
116 | Y>C | Intellectual disability, autosomal dominant 27 IDDMOH; decreases transcriptional activity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1572216329 RCV000995881 |
118 | Y>S | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV000623196 RCV002466550 CA345866535 rs749901648 RCV001569723 |
119 | R>P | Coffin-Siris syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
| VAR_088058 | 120 | P>H | IDDMOH; decreased function in positive regulation of DNA-templated transcription; compared to the wild-type, the mutant induces a reduced GDF5 promoter activation using an in vitro reporter system [UniProt] | Yes | UniProt |
|
rs1665662335 RCV001251188 |
120 | P>L | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_088059 | 142 | A>G | IDDMOH; uncertain significance; no effect on nuclear localization [UniProt] | Yes | UniProt |
| VAR_088060 | 176 | A>E | IDDMOH; uncertain significance [UniProt] | Yes | UniProt |
|
rs1297634063 RCV001291810 |
180 | A>T | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs780122780 RCV001331021 |
234 | E>* | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_088061 | 234 | E>del | IDDMOH [UniProt] | Yes | UniProt |
|
rs745675837 RCV001335094 |
263 | P>T | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
CA345867460 RCV000623171 rs1553327954 RCV003224877 |
264 | S>* | Intellectual disability, autosomal dominant 27 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| VAR_088062 | 274 | K>del | IDDMOH [UniProt] | Yes | UniProt |
|
rs1388073236 RCV001257281 |
295 | D>E | Pituitary stalk interruption syndrome [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs991368727 RCV000986587 |
370 | S>F | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
|
rs1665683367 RCV001335093 |
383 | G>C | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA658795673 rs1553328057 RCV000623189 |
389 | N>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
|
RCV000824995 rs1572217107 |
406 | S>missing | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000414848 rs1057518672 CA16043657 |
429 | W>* | Intellectual disability, autosomal dominant 27 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| TCGA novel | 3 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1337380916 | 4 | Q>* | No | gnomAD | |
| rs1273091935 | 5 | A>E | No | gnomAD | |
| rs755780929 | 5 | A>T | No |
ExAC gnomAD |
|
| TCGA novel | 10 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756768286 | 11 | E>* | No |
ExAC gnomAD |
|
|
COSM461005 rs369950584 |
11 | E>D | cervix Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs890630265 | 12 | S>I | No |
TOPMed gnomAD |
|
| rs890630265 | 12 | S>N | No |
TOPMed gnomAD |
|
| rs772578286 | 12 | S>R | No |
ExAC gnomAD |
|
| TCGA novel | 13 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1008105213 | 13 | N>K | No | Ensembl | |
| rs1017699278 | 15 | P>H | No |
TOPMed gnomAD |
|
| rs1164202412 | 15 | P>S | No | gnomAD | |
|
rs1411163388 COSM5082155 |
16 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs149438305 | 17 | E>Q | No |
ESP ExAC gnomAD |
|
|
COSM1266485 rs768889757 |
18 | A>V | oesophagus [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs773428827 | 22 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs765299842 | 22 | E>Q | No |
ExAC gnomAD |
|
| rs1572216193 | 23 | E>D | No | Ensembl | |
| COSM4094943 | 25 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1363221380 | 27 | M>I | No |
TOPMed gnomAD |
|
| rs1481974602 | 27 | M>T | No | TOPMed | |
| rs959309748 | 29 | C>F | No |
TOPMed gnomAD |
|
| rs959309748 | 29 | C>Y | No |
TOPMed gnomAD |
|
| rs1572216212 | 32 | V>G | No | Ensembl | |
| rs991720486 | 32 | V>L | No | TOPMed | |
| rs1572216224 | 34 | L>P | No | Ensembl | |
| rs752503007 | 35 | D>E | No |
ExAC gnomAD |
|
| rs1316753803 | 35 | D>H | No | TOPMed | |
| rs1242668001 | 37 | S>N | No | gnomAD | |
| rs1476324230 | 38 | D>G | No | gnomAD | |
| COSM1227167 | 45 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4434777 | 46 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 46 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 49 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 50 | K>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1021938 | 53 | M>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4094944 | 57 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758858972 | 59 | W>C | No |
ExAC gnomAD |
|
| rs1305912185 | 62 | I>M | No | gnomAD | |
| TCGA novel | 63 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1558373180 | 63 | E>Q | No | Ensembl | |
|
RCV000483495 CA16617743 rs1064794702 |
64 | R>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
| TCGA novel | 66 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1284756290 | 67 | I>V | No | gnomAD | |
| COSM4830579 | 70 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4846715 | 72 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 72 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1448489215 | 72 | P>S | No | gnomAD | |
| TCGA novel | 78 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1665660680 RCV001192496 |
79 | I>L | No |
ClinVar dbSNP |
|
| rs202211730 | 86 | R>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 89 | M>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770070167 | 90 | L>M | No |
ExAC gnomAD |
|
| TCGA novel | 95 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763158621 | 96 | I>T | No |
ExAC gnomAD |
|
| rs1064794628 | 100 | R>Q | No | gnomAD | |
| COSM3733645 | 100 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 104 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 105 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1665661722 RCV001267933 |
108 | K>T | No |
ClinVar dbSNP |
|
| rs1182560635 | 110 | M>I | No | TOPMed | |
| TCGA novel | 111 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 111 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs866692324 | 111 | A>V | No | Ensembl | |
| rs1280356263 | 114 | P>H | No | gnomAD | |
| TCGA novel | 114 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1408741 rs1221789039 |
115 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA gnomAD |
| COSM443056 | 115 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 117 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749901648 | 119 | R>Q | No |
ExAC gnomAD |
|
| rs757949814 | 120 | P>T | No |
ExAC gnomAD |
|
| TCGA novel | 121 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4094947 | 121 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 124 | P>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1035983892 | 125 | K>I | No | Ensembl | |
| rs1182148169 | 126 | M>I | No | gnomAD | |
| rs1485846444 | 126 | M>T | No | TOPMed | |
| rs1572216350 | 127 | D>A | No | Ensembl | |
| rs755512766 | 127 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1471365952 | 128 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs573393747 | 128 | P>S | No |
1000Genomes ExAC TOPMed |
|
| rs573393747 | 128 | P>T | No |
1000Genomes ExAC TOPMed |
|
| rs1411909990 | 129 | S>W | No | gnomAD | |
| rs1462477218 | 130 | A>T | No | gnomAD | |
| rs1167697596 | 130 | A>V | No | gnomAD | |
| rs199680382 | 131 | K>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1386272998 | 132 | P>L | No | gnomAD | |
| rs991709110 | 133 | S>G | No | TOPMed | |
| rs562403473 | 133 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs374256122 | 134 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs374256122 | 134 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs749358189 | 134 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs374256122 | 134 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1271177335 | 140 | K>E | No | gnomAD | |
| rs1355052745 | 141 | S>G | No | gnomAD | |
| rs1252271637 | 142 | A>V | No | gnomAD | |
| TCGA novel | 143 | A>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1475737480 | 144 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1251597664 | 144 | G>C | No | gnomAD | |
| rs1251597664 | 144 | G>S | No | gnomAD | |
| rs761704672 | 146 | G>A | No |
ExAC gnomAD |
|
| rs761704672 | 146 | G>D | No |
ExAC gnomAD |
|
|
RCV000426910 rs749379484 CA1517836 |
147 | G>missing | No |
ClinGen ClinVar dbSNP |
|
| rs775298170 | 147 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1398697866 | 148 | G>A | No |
TOPMed gnomAD |
|
| rs1398697866 | 148 | G>E | No |
TOPMed gnomAD |
|
| rs764844599 | 148 | G>R | No |
ExAC gnomAD |
|
| rs943373465 | 149 | S>R | No |
TOPMed gnomAD |
|
| rs1291365883 | 150 | A>E | No |
TOPMed gnomAD |
|
| rs750131174 | 150 | A>S | No |
ExAC gnomAD |
|
| rs750131174 | 150 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1227551760 | 151 | G>S | No | gnomAD | |
|
RCV000722661 rs774530705 |
153 | G>missing | No |
ClinVar dbSNP |
|
| TCGA novel | 154 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1038912868 | 156 | G>C | No |
TOPMed gnomAD |
|
| rs1190433874 | 157 | A>T | No | gnomAD | |
| rs1254711016 | 158 | K>E | No | gnomAD | |
| rs865924581 | 160 | S>P | No | Ensembl | |
| rs781738124 | 162 | G>S | No |
ExAC gnomAD |
|
| rs1374927340 | 164 | S>N | No | gnomAD | |
| rs1474457843 | 165 | K>Q | No | gnomAD | |
| rs752975676 | 166 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs946669836 | 167 | C>S | No | TOPMed | |
| rs1376216339 | 170 | L>I | No | gnomAD | |
|
CA345866868 rs1553327863 RCV000512828 |
171 | K>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs902373698 | 174 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
COSM5036609 rs1436393730 |
174 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1340253906 | 176 | A>T | No | gnomAD | |
| rs1384818454 | 178 | A>P | No | TOPMed | |
| rs1035189329 | 180 | A>E | No |
TOPMed gnomAD |
|
| rs1420816592 | 181 | G>S | No | TOPMed | |
| rs1349218134 | 181 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1208615367 | 182 | A>G | No | gnomAD | |
| rs1478438158 | 182 | A>S | No | TOPMed | |
| rs894770529 | 183 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs894770529 | 183 | G>S | No |
TOPMed gnomAD |
|
| rs1268368634 | 184 | K>R | No | TOPMed | |
| rs1195435511 | 185 | A>E | No | TOPMed | |
| rs1195435511 | 185 | A>V | No | TOPMed | |
| rs1208248317 | 186 | A>G | No | gnomAD | |
| rs1462673426 | 187 | Q>H | No |
TOPMed gnomAD |
|
| rs1260086974 | 189 | G>R | No | TOPMed | |
| rs377528494 | 190 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1463602758 | 192 | G>V | No | gnomAD | |
| rs745988357 | 193 | G>D | No |
ExAC gnomAD |
|
| rs779081464 | 193 | G>S | No |
ExAC gnomAD |
|
| rs1171980257 | 194 | A>T | No |
TOPMed gnomAD |
|
| rs1454109830 | 195 | G>D | No |
TOPMed gnomAD |
|
| rs1376928154 | 195 | G>S | No | gnomAD | |
| rs544581532 | 196 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776676775 | 196 | D>N | No |
ExAC gnomAD |
|
| rs776676775 | 196 | D>Y | No |
ExAC gnomAD |
|
| rs564494780 | 197 | D>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1227165 rs564494780 |
197 | D>N | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs1057518187 CA16042463 RCV000414399 |
198 | Y>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs766041973 | 198 | Y>C | No |
ExAC gnomAD |
|
| TCGA novel | 198 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773695032 | 199 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1009563168 | 200 | L>R | No | Ensembl | |
| rs753307477 | 203 | L>M | No |
ExAC gnomAD |
|
| rs753307477 | 203 | L>V | No |
ExAC gnomAD |
|
| rs756544888 | 204 | R>C | No |
ExAC gnomAD |
|
| rs764489337 | 204 | R>L | No |
ExAC gnomAD |
|
| rs764489337 | 204 | R>P | No |
ExAC gnomAD |
|
| rs1479245018 | 205 | V>G | No | TOPMed | |
| rs1208628926 | 205 | V>M | No | gnomAD | |
| TCGA novel | 206 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1468575248 | 207 | G>R | No | gnomAD | |
| rs1469878578 | 208 | S>L | No | gnomAD | |
| rs1189755341 | 209 | G>D | No | gnomAD | |
| rs748010810 | 210 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs748010810 | 210 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1456675614 | 212 | G>D | No | gnomAD | |
| rs1409457496 | 212 | G>S | No | gnomAD | |
| rs770541398 | 213 | A>E | No |
ExAC gnomAD |
|
| rs770541398 | 213 | A>G | No |
ExAC gnomAD |
|
| rs1438003053 | 214 | G>D | No | gnomAD | |
| rs1373555541 | 216 | T>M | No | gnomAD | |
| rs1309670314 | 217 | V>A | No | gnomAD | |
| TCGA novel | 217 | V>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758958162 | 220 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1228396860 | 223 | D>N | No |
TOPMed gnomAD |
|
| rs1558373704 | 225 | D>E | No | Ensembl | |
| rs767125292 | 225 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1337057292 | 226 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs761209390 | 227 | D>E | No |
ExAC gnomAD |
|
| rs774871082 | 227 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs774871082 | 227 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs764724459 | 228 | D>E | No |
ExAC gnomAD |
|
| rs1572216621 | 228 | D>H | No | Ensembl | |
| rs757741965 | 230 | D>E | No |
ExAC gnomAD |
|
| rs1179136145 | 230 | D>N | No | gnomAD | |
| rs754220187 | 230 | D>V | No |
ExAC gnomAD |
|
| rs750721557 | 231 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs765361062 | 231 | D>G | No |
ExAC gnomAD |
|
| rs1403124561 | 232 | D>N | No |
TOPMed gnomAD |
|
| rs1354586764 | 233 | D>E | No |
TOPMed gnomAD |
|
| rs1042297449 | 233 | D>H | No |
TOPMed gnomAD |
|
| rs1042297449 | 233 | D>N | No |
TOPMed gnomAD |
|
| rs751660162 | 234 | E>G | No |
ExAC TOPMed gnomAD |
|
| COSM1720521 | 234 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780122780 | 234 | E>Q | No |
ExAC gnomAD |
|
| rs756004827 | 235 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1226706343 | 237 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
CA1517903 RCV000500849 RCV001764496 rs140772793 |
237 | L>V | No |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM6091694 rs1256714607 COSM575626 |
238 | Q>H | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed |
| COSM6158313 | 238 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778428159 | 240 | K>R | No |
ExAC TOPMed |
|
| rs745460988 | 241 | Q>R | No |
ExAC gnomAD |
|
| rs1206958743 | 242 | E>Q | No | gnomAD | |
| rs771639613 | 243 | P>A | No |
ExAC gnomAD |
|
| rs769259145 | 244 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs760297024 | 244 | D>N | No |
ExAC gnomAD |
|
| rs1471796361 | 245 | E>K | No |
TOPMed gnomAD |
|
| rs1471796361 | 245 | E>Q | No |
TOPMed gnomAD |
|
| rs755851269 | 247 | D>Y | No |
ExAC gnomAD |
|
| rs765568135 | 248 | E>K | No |
ExAC gnomAD |
|
| rs765568135 | 248 | E>Q | No |
ExAC gnomAD |
|
| rs1166348174 | 249 | E>K | No | gnomAD | |
| rs1355135739 | 250 | P>T | No | TOPMed | |
| rs1313818654 | 251 | P>Q | No | gnomAD | |
| rs1429550919 | 251 | P>T | No | gnomAD | |
| rs1362764728 | 254 | Q>K | No | gnomAD | |
| rs1281925996 | 255 | L>F | No | TOPMed | |
| rs868826903 | 256 | L>P | No | Ensembl | |
| rs865777669 | 257 | Q>P | No | Ensembl | |
| rs868349180 | 258 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs755100945 | 258 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs866041767 | 259 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs757148387 | 260 | G>E | No |
ExAC gnomAD |
|
| rs1186080588 | 261 | Q>* | No |
TOPMed gnomAD |
|
| rs1186080588 | 261 | Q>K | No |
TOPMed gnomAD |
|
| rs779898240 | 263 | P>L | No |
ExAC gnomAD |
|
| rs745675837 | 263 | P>S | No |
ExAC gnomAD |
|
| rs762321557 | 267 | L>Q | No |
ExAC gnomAD |
|
| rs770223329 | 269 | R>H | No |
ExAC gnomAD |
|
| rs1437389899 | 270 | Y>H | No | gnomAD | |
| rs1364504443 | 271 | N>H | No | gnomAD | |
| rs763266934 | 272 | V>L | No |
ExAC gnomAD |
|
| rs1276818965 | 273 | A>P | No |
TOPMed gnomAD |
|
| rs1276818965 | 273 | A>S | No |
TOPMed gnomAD |
|
| rs1218359229 | 276 | P>S | No | gnomAD | |
| rs1284912385 | 280 | T>M | No | gnomAD | |
| TCGA novel | 281 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1246927388 | 282 | S>N | No | gnomAD | |
| TCGA novel | 285 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs943565885 | 287 | S>F | No | Ensembl | |
| rs971298974 | 287 | S>P | No | gnomAD | |
| rs971298974 | 287 | S>T | No | gnomAD | |
| rs765121971 | 291 | A>E | No |
ExAC gnomAD |
|
| rs765121971 | 291 | A>G | No |
ExAC gnomAD |
|
| rs1441654871 | 296 | E>* | No | gnomAD | |
| rs779845221 | 297 | V>A | No |
ExAC gnomAD |
|
| rs1435531266 | 298 | R>Q | No | gnomAD | |
| rs780798415 | 301 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs780798415 | 301 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs780798415 | 301 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1267359699 | 302 | T>I | No | gnomAD | |
| rs1252447458 | 303 | S>L | No | gnomAD | |
| rs1334919509 | 303 | S>T | No | gnomAD | |
| rs1252447458 | 303 | S>W | No | gnomAD | |
| rs1287329032 | 304 | G>C | No | gnomAD | |
| rs773641572 | 305 | A>P | No |
ExAC gnomAD |
|
| rs773641572 | 305 | A>S | No |
ExAC gnomAD |
|
| rs1180668384 | 307 | G>C | No |
TOPMed gnomAD |
|
| rs1180668384 | 307 | G>R | No |
TOPMed gnomAD |
|
| rs1476800720 | 307 | G>V | No | TOPMed | |
| rs759699166 | 308 | G>S | No |
ExAC gnomAD |
|
| rs767752007 | 309 | S>C | No |
ExAC gnomAD |
|
| rs1388644510 | 309 | S>N | No |
TOPMed gnomAD |
|
| rs775602980 | 310 | R>H | No |
ExAC gnomAD |
|
| rs760713336 | 311 | L>H | No |
ExAC gnomAD |
|
| rs1287678940 | 318 | I>M | No | gnomAD | |
| rs1355953682 | 320 | K>Q | No | gnomAD | |
| rs1246870502 | 321 | Q>* | No | gnomAD | |
|
rs1553327990 CA658795672 RCV000627527 |
322 | H>missing | No |
ClinGen ClinVar dbSNP |
|
| rs1003136889 | 322 | H>P | No | Ensembl | |
| TCGA novel | 322 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1355029618 | 323 | P>R | No | gnomAD | |
| rs1276263281 | 324 | P>L | No | gnomAD | |
| rs747671372 | 330 | A>V | No |
ExAC gnomAD |
|
| rs1481369496 | 331 | L>R | No |
TOPMed gnomAD |
|
| rs1472474340 | 332 | S>L | No |
TOPMed gnomAD |
|
| rs749826506 | 334 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs774743661 | 335 | S>C | No |
ExAC gnomAD |
|
| rs1375803449 | 336 | S>L | No | gnomAD | |
|
RCV001268074 rs1665680497 |
337 | R>G | No |
ClinVar dbSNP |
|
| rs772309412 | 337 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs772309412 | 337 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1339382294 | 338 | S>L | No | gnomAD | |
| rs1227439657 | 339 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1339574821 | 343 | S>W | No | gnomAD | |
| rs1003926124 | 344 | S>F | No | Ensembl | |
| rs775521809 | 344 | S>T | No |
ExAC gnomAD |
|
| rs1278356774 | 345 | S>G | No | Ensembl | |
| rs1274427173 | 345 | S>N | No | gnomAD | |
| rs1483782583 | 345 | S>R | No |
TOPMed gnomAD |
|
| rs1202768939 | 347 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs760818536 | 347 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1409527492 | 350 | S>R | No | gnomAD | |
| rs776762311 | 352 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs776762311 | 352 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs776762311 | 352 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs766389339 | 352 | G>V | No |
ExAC gnomAD |
|
| rs1447931988 | 353 | S>N | No | gnomAD | |
|
COSM1614980 rs1395519132 |
355 | G>D | liver [Cosmic] | No |
cosmic curated gnomAD |
| rs751432316 | 355 | G>S | No |
ExAC gnomAD |
|
| rs767290813 | 357 | D>H | No |
ExAC gnomAD |
|
|
rs767290813 COSM3582376 |
357 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1362366101 | 362 | M>V | No | gnomAD | |
| rs1403614666 | 363 | F>L | No | gnomAD | |
| rs1558374095 | 364 | D>Y | No | Ensembl | |
| rs974822756 | 366 | S>C | No | TOPMed | |
| rs1313151836 | 369 | F>L | No | gnomAD | |
| rs1235827793 | 372 | S>R | No | gnomAD | |
| rs1344979183 | 374 | H>R | No | gnomAD | |
| rs1449180406 | 375 | S>N | No | TOPMed | |
| rs1377727959 | 375 | S>R | No | TOPMed | |
| rs1022798363 | 376 | A>G | No | Ensembl | |
| rs1209872000 | 376 | A>S | No | gnomAD | |
| rs762687158 | 377 | S>R | No |
TOPMed gnomAD |
|
| rs757798729 | 380 | Q>L | No |
ExAC gnomAD |
|
| rs1188178837 | 381 | L>P | No |
TOPMed gnomAD |
|
| rs1188178837 | 381 | L>R | No |
TOPMed gnomAD |
|
| rs924321972 | 382 | G>R | No | Ensembl | |
| rs1553328038 | 383 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1173636535 | 383 | G>D | No | gnomAD | |
|
CA16042434 rs1057518282 RCV000412808 |
385 | A>missing | No |
ClinGen ClinVar dbSNP |
|
| rs764036560 | 385 | A>E | No | Ensembl | |
| TCGA novel | 385 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1454287063 | 386 | A>G | No | gnomAD | |
| rs1572217038 | 389 | N>D | No | Ensembl | |
| TCGA novel | 391 | S>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 393 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1572217054 | 395 | V>G | No | Ensembl | |
| rs1572217069 | 396 | D>G | No | Ensembl | |
| rs1189353231 | 397 | K>T | No | gnomAD | |
| COSM1565527 | 398 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1482580454 | 400 | D>N | No | TOPMed | |
| rs1229609930 | 401 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1212106612 | 402 | F>L | No | TOPMed | |
| TCGA novel | 403 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1208145337 | 403 | S>R | No |
TOPMed gnomAD |
|
| TCGA novel | 406 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1189662451 | 410 | H>N | No | gnomAD | |
| rs1428529342 | 411 | F>I | No | gnomAD | |
| COSM35844 | 412 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1266498989 | 412 | E>K | No | gnomAD | |
| COSM1408744 | 416 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1317674231 | 417 | C>S | No | TOPMed | |
| TCGA novel | 419 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1021941 rs1409607021 |
423 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 424 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 425 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761946808 | 426 | A>G | No |
ExAC gnomAD |
|
| TCGA novel | 426 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774373152 | 428 | D>Y | No |
ExAC gnomAD |
|
| rs1572217136 | 429 | W>* | No | Ensembl | |
| rs1394318840 | 429 | W>R | No | TOPMed | |
| rs759409441 | 431 | E>G | No |
ExAC gnomAD |
|
| rs767281459 | 432 | A>G | No | ExAC | |
| rs1390480045 | 432 | A>T | No | gnomAD | |
| rs767281459 | 432 | A>V | No | ExAC | |
| rs1572217159 | 433 | N>D | No | Ensembl | |
| TCGA novel | 434 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1572217168 | 438 | V>G | No | Ensembl | |
| TCGA novel | 441 | Y>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
1 associated diseases with P35716
[MIM: 615866]: Intellectual developmental disorder with microcephaly and with or without ocular malformations or hypogonadotropic hypogonadism (IDDMOH)
An autosomal dominant disorder characterized by developmental delay, impaired intellectual development and microcephaly. Affected individuals may also have oculomotor apraxia, ocular malformations including coloboma, lens abnormalities and microphthalmia, and hypogonadotropic hypogonadism. Some patients may have finger clinodactyly and hypoplastic distal phalanges with nail hypoplasia, especially of the fifth digits. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant disorder characterized by developmental delay, impaired intellectual development and microcephaly. Affected individuals may also have oculomotor apraxia, ocular malformations including coloboma, lens abnormalities and microphthalmia, and hypogonadotropic hypogonadism. Some patients may have finger clinodactyly and hypoplastic distal phalanges with nail hypoplasia, especially of the fifth digits. . Note=The disease is caused by variants affecting the gene represented in this entry.
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR10270 | SOX TRANSCRIPTION FACTOR |
| PANTHER Subfamily | PTHR10270:SF113 | TRANSCRIPTION FACTOR SOX-11 |
| PANTHER Protein Class |
DNA-binding transcription factor
HMG box transcription factor |
|
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
39 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure morphogenesis | The process in which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form. |
| closure of optic fissure | The closure of the temporary ventral gap in the optic cup that contributes to its shaping. |
| cornea development in camera-type eye | The progression of the cornea over time, from its formation to the mature structure. The cornea is the transparent structure that covers the anterior of the eye. |
| embryonic digestive tract morphogenesis | The process in which the anatomical structures of the digestive tract are generated and organized during embryonic development. The digestive tract is the anatomical structure through which food passes and is processed. |
| embryonic skeletal system morphogenesis | The process in which the anatomical structures of the skeleton are generated and organized during the embryonic phase. |
| eyelid development in camera-type eye | The progression of the eyelid in a camera-type eye from its formation to the mature state. The eyelid is a membranous cover that helps protect and lubricate the eye. |
| glial cell proliferation | The multiplication or reproduction of glial cells by cell division, resulting in the expansion of their population. Glial cells exist throughout the nervous system, and include Schwann cells, astrocytes, and oligodendrocytes among others. |
| hard palate development | The biological process whose specific outcome is the progression of the hard palate from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. The hard palate is the anterior portion of the palate consisting of bone and mucous membranes. |
| kidney development | The process whose specific outcome is the progression of the kidney over time, from its formation to the mature structure. The kidney is an organ that filters the blood and/or excretes the end products of body metabolism in the form of urine. |
| lens morphogenesis in camera-type eye | The process in which the anatomical structures of the lens are generated and organized. The lens is a transparent structure in the eye through which light is focused onto the retina. An example of this process is found in Mus musculus. |
| lung morphogenesis | The process in which the anatomical structures of the lung are generated and organized. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of glial cell proliferation | Any process that stops or decreases the rate or extent of glial cell proliferation. |
| negative regulation of lymphocyte proliferation | Any process that stops, prevents or reduces the rate or extent of lymphocyte proliferation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transcription regulatory region DNA binding | Any process that stops, prevents or reduces the frequency, rate or extent of transcription regulatory region DNA binding. |
| nervous system development | The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state. |
| neuroepithelial cell differentiation | The process in which epiblast cells acquire specialized features of neuroepithelial cells. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| noradrenergic neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of an noradrenergic neuron, a neuron that secretes noradrenaline. |
| oligodendrocyte development | The process aimed at the progression of an oligodendrocyte over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell. An oligodendrocyte is a type of glial cell involved in myelinating the axons in the central nervous system. |
| outflow tract morphogenesis | The process in which the anatomical structures of the outflow tract are generated and organized. The outflow tract is the portion of the heart through which blood flows into the arteries. |
| positive regulation of BMP signaling pathway | Any process that activates or increases the frequency, rate or extent of BMP signaling pathway activity. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of hippo signaling | Any process that activates or increases the frequency, rate or extent of hippo signaling. |
| positive regulation of hormone secretion | Any process that activates or increases the frequency, rate or extent of the regulated release of a hormone from a cell. |
| positive regulation of lens epithelial cell proliferation | Any process that activates or increases the frequency, rate or extent of lens epithelial cell proliferation. |
| positive regulation of neurogenesis | Any process that activates or increases the frequency, rate or extent of neurogenesis, the generation of cells within the nervous system. |
| positive regulation of neuron differentiation | Any process that activates or increases the frequency, rate or extent of neuron differentiation. |
| positive regulation of ossification | Any process that activates or increases the frequency, rate or extent of ossification, the formation of bone or of a bony substance or the conversion of fibrous tissue or of cartilage into bone or a bony substance. |
| positive regulation of osteoblast differentiation | Any process that activates or increases the frequency, rate or extent of osteoblast differentiation. |
| positive regulation of stem cell proliferation | Any process that activates or increases the frequency, rate or extent of stem cell proliferation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of transforming growth factor beta receptor signaling pathway | Any process that modulates the frequency, rate or extent of activity of any TGF-beta receptor signaling pathway. |
| skeletal muscle cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a skeletal muscle cell, a somatic cell located in skeletal muscle. |
| soft palate development | The biological process whose specific outcome is the progression of the soft palate from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. The soft palate is the posterior portion of the palate extending from the posterior edge of the hard palate. |
| spinal cord development | The process whose specific outcome is the progression of the spinal cord over time, from its formation to the mature structure. The spinal cord primarily conducts sensory and motor nerve impulses between the brain and the peripheral nervous tissues. |
| sympathetic nervous system development | The process whose specific outcome is the progression of the sympathetic nervous system over time, from its formation to the mature structure. The sympathetic nervous system is one of the two divisions of the vertebrate autonomic nervous system (the other being the parasympathetic nervous system). The sympathetic preganglionic neurons have their cell bodies in the thoracic and lumbar regions of the spinal cord and connect to the paravertebral chain of sympathetic ganglia. Innervate heart and blood vessels, sweat glands, viscera and the adrenal medulla. Most sympathetic neurons, but not all, use noradrenaline as a post-ganglionic neurotransmitter. |
| ventricular septum morphogenesis | The developmental process in which a ventricular septum is generated and organized. A ventricular septum is an anatomical structure that separates the lower chambers (ventricles) of the heart from one another. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P48435 | SOX11 | Transcription factor SOX-11 | Gallus gallus (Chicken) | SS |
| P48436 | SOX9 | Transcription factor SOX-9 | Homo sapiens (Human) | PR |
| O94993 | SOX30 | Transcription factor SOX-30 | Homo sapiens (Human) | PR |
| O15370 | SOX12 | Transcription factor SOX-12 | Homo sapiens (Human) | SS |
| Q04888 | Sox10 | Transcription factor SOX-10 | Mus musculus (Mouse) | PR |
| Q05738 | Sry | Sex-determining region Y protein | Mus musculus (Mouse) | SS |
| Q04890 | Sox12 | Transcription factor SOX-12 | Mus musculus (Mouse) | SS |
| Q04886 | Sox8 | Transcription factor SOX-8 | Mus musculus (Mouse) | SS |
| Q04887 | Sox9 | Transcription factor SOX-9 | Mus musculus (Mouse) | PR |
| Q7M6Y2 | Sox11 | Transcription factor SOX-11 | Mus musculus (Mouse) | SS |
| P0C1G9 | Sox11 | Transcription factor SOX-11 | Rattus norvegicus (Rat) | SS |
| Q8T3B9 | sem-2 | Transcription factor sem-2 | Caenorhabditis elegans | SS |
| Q6GLH8 | sox17b.2 | Transcription factor Sox-17-beta.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q66JF1 | sox11 | Transcription factor Sox-11 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVQQAESLEA | ESNLPREALD | TEEGEFMACS | PVALDESDPD | WCKTASGHIK | RPMNAFMVWS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KIERRKIMEQ | SPDMHNAEIS | KRLGKRWKML | KDSEKIPFIR | EAERLRLKHM | ADYPDYKYRP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RKKPKMDPSA | KPSASQSPEK | SAAGGGGGSA | GGGAGGAKTS | KGSSKKCGKL | KAPAAAGAKA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GAGKAAQSGD | YGGAGDDYVL | GSLRVSGSGG | GGAGKTVKCV | FLDEDDDDDD | DDDELQLQIK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QEPDEEDEEP | PHQQLLQPPG | QQPSQLLRRY | NVAKVPASPT | LSSSAESPEG | ASLYDEVRAG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ATSGAGGGSR | LYYSFKNITK | QHPPPLAQPA | LSPASSRSVS | TSSSSSSGSS | SGSSGEDADD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LMFDLSLNFS | QSAHSASEQQ | LGGGAAAGNL | SLSLVDKDLD | SFSEGSLGSH | FEFPDYCTPE |
| 430 | 440 | ||||
| LSEMIAGDWL | EANFSDLVFT | Y |