P35283
Gene name |
Rab12 |
Protein name |
Ras-related protein Rab-12 |
Names |
Rab-13 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:19328 |
EC number |
|
Protein Class |
|
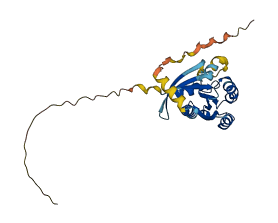
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P35283
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P35283-F1 | Predicted | AlphaFoldDB |
No variants for P35283
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P35283 | |||||
No associated diseases with P35283
1 regional properties for P35283
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 43 - 196 | IPR005225 |
Functions
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| autophagosome | A double-membrane-bounded compartment that engulfs endogenous cellular material as well as invading microorganisms to target them to the lytic vacuole/lysosome for degradation as part of macroautophagy. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| insulin-responsive compartment | A small membrane-bounded vesicle that releases its contents by exocytosis in response to insulin stimulation; the contents are enriched in GLUT4, IRAP and VAMP2. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| lysosome | A small lytic vacuole that has cell cycle-independent morphology found in most animal cells and that contains a variety of hydrolases, most of which have their maximal activities in the pH range 5-6. The contained enzymes display latency if properly isolated. About 40 different lysosomal hydrolases are known and lysosomes have a great variety of morphologies and functions. |
| phagocytic vesicle | A membrane-bounded intracellular vesicle that arises from the ingestion of particulate material by phagocytosis. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
| recycling endosome membrane | The lipid bilayer surrounding a recycling endosome. |
| secretory granule | A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cellular response to interferon-gamma | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interferon-gamma stimulus. Interferon gamma is the only member of the type II interferon found so far. |
| endosome to lysosome transport | The directed movement of substances from endosomes to lysosomes. |
| positive regulation of macroautophagy | Any process, such as recognition of nutrient depletion, that activates or increases the rate of macroautophagy to bring cytosolic macromolecules to the vacuole/lysosome for degradation. |
| protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein secretion | The controlled release of proteins from a cell. |
| Rab protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rab family of proteins switching to a GTP-bound active state. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| vesicle docking involved in exocytosis | The initial attachment of a vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the vesicle and the target membrane, that contributes to exocytosis. |
61 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P07560 | SEC4 | Ras-related protein SEC4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q2HJI8 | RAB8B | Ras-related protein Rab-8B | Bos taurus (Bovine) | PR |
| A4FV54 | RAB8A | Ras-related protein Rab-8A | Bos taurus (Bovine) | PR |
| Q1RMR4 | RAB15 | Ras-related protein Rab-15 | Bos taurus (Bovine) | PR |
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| Q5F470 | RAB8A | Ras-related protein Rab-8A | Gallus gallus (Chicken) | PR |
| P51152 | RAB12 | Ras-related protein Rab-12 | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| P0C0E4 | RAB40AL | Ras-related protein Rab-40A-like | Homo sapiens (Human) | PR |
| P61006 | RAB8A | Ras-related protein Rab-8A | Homo sapiens (Human) | PR |
| Q92930 | RAB8B | Ras-related protein Rab-8B | Homo sapiens (Human) | PR |
| Q8WXH6 | RAB40A | Ras-related protein Rab-40A | Homo sapiens (Human) | PR |
| Q96S21 | RAB40C | Ras-related protein Rab-40C | Homo sapiens (Human) | PR |
| Q12829 | RAB40B | Ras-related protein Rab-40B | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| Q8CB87 | Rab44 | Ras-related protein Rab-44 | Mus musculus (Mouse) | PR |
| P55258 | Rab8a | Ras-related protein Rab-8A | Mus musculus (Mouse) | PR |
| P61028 | Rab8b | Ras-related protein Rab-8B | Mus musculus (Mouse) | PR |
| Q8VHQ4 | Rab40c | Ras-related protein Rab-40C | Mus musculus (Mouse) | PR |
| Q9DD03 | Rab13 | Ras-related protein Rab-13 | Mus musculus (Mouse) | PR |
| Q8VHP8 | Rab40b | Ras-related protein Rab-40B | Mus musculus (Mouse) | PR |
| Q8K386 | Rab15 | Ras-related protein Rab-15 | Mus musculus (Mouse) | PR |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| P35289 | Rab15 | Ras-related protein Rab-15 | Rattus norvegicus (Rat) | PR |
| P35280 | Rab8a | Ras-related protein Rab-8A | Rattus norvegicus (Rat) | PR |
| P70550 | Rab8b | Ras-related protein Rab-8B | Rattus norvegicus (Rat) | PR |
| P35281 | Rab10 | Ras-related protein Rab-10 | Rattus norvegicus (Rat) | PR |
| P35284 | Rab12 | Ras-related protein Rab-12 | Rattus norvegicus (Rat) | PR |
| O24466 | RABE1A | Ras-related protein RABE1a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SF91 | RABE1E | Ras-related protein RABE1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LZD4 | RABE1D | Ras-related protein RABE1d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDPSAALHRR | PAGGSLGAVS | PALSGGQARR | RKQPPRPADF | KLQVIIIGSR | GVGKTSLMER |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FTDDTFCEAC | KSTVGVDFKI | KTVELRGKKI | RLQIWDTAGQ | ERFNSITSAY | YRSAKGIILV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YDITKKETFD | DLPKWMKMID | KYASEDAELL | LVGNKLDCET | DREISRQQGE | KFAQQITGMR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FCEASAKDNF | NVDEIFLKLV | DDILKKMPLD | VLRNELSNSI | LSLQPEPEIP | PELPPPRPHV |
| RCC |