P34931
Gene name |
HSPA1L |
Protein name |
Heat shock 70 kDa protein 1-like |
Names |
Heat shock 70 kDa protein 1L , Heat shock 70 kDa protein 1-Hom , HSP70-Hom , Heat shock protein family A member 1L |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:3305 |
EC number |
|
Protein Class |
|
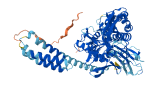
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
227-264 (Nucleolar targeting region) |
Relief mechanism |
Others |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P34931
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3GDQ | X-ray | 180 A | A | 1-386 | PDB |
| AF-P34931-F1 | Predicted | AlphaFoldDB |
704 variants for P34931
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA136890474 RCV001782884 rs368138379 |
77 | G>S | Inflammatory bowel disease 1 [ClinVar] | Yes |
ClinGen ClinVar ESP TOPMed dbSNP gnomAD |
|
RCV001782885 rs750447828 CA3724060 |
172 | L>missing | Inflammatory bowel disease 1 [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
|
CA3724008 RCV001782883 rs139868987 |
267 | T>I | Inflammatory bowel disease 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_025842 RCV001782882 CA3724007 RCV003430979 rs34620296 |
268 | A>T | Inflammatory bowel disease 1 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000856580 rs2227956 |
493 | T>K | Chronic obstructive pulmonary disease [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA3723860 RCV001692092 rs2227955 RCV001782881 VAR_025845 |
558 | E>A | Inflammatory bowel disease 1 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1815457330 | 2 | A>P | No | TOPMed | |
| rs140839023 | 3 | T>A | No | ESP | |
| rs147186823 | 4 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs372378700 | 4 | A>T | No |
ESP ExAC gnomAD |
|
| rs768906825 | 5 | K>N | No |
ExAC gnomAD |
|
| rs749753182 | 6 | G>* | No |
ExAC gnomAD |
|
| rs1815455712 | 6 | G>V | No | Ensembl | |
|
VAR_025841 rs9469057 |
8 | A>P | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs9469057 | 8 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 8 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139193421 | 9 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs780700552 | 9 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs151059204 | 9 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777589589 | 10 | G>S | No |
ExAC gnomAD |
|
| rs1265682797 | 12 | D>H | No |
TOPMed gnomAD |
|
|
COSM1077719 rs1265682797 |
12 | D>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1265682797 | 12 | D>Y | No |
TOPMed gnomAD |
|
| COSM3861839 | 13 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752662698 | 14 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs765307007 | 17 | Y>* | No |
ExAC gnomAD |
|
| rs1007813910 | 18 | S>C | No |
TOPMed gnomAD |
|
| rs1562343316 | 18 | S>P | No | Ensembl | |
| rs760834707 | 19 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1815451496 | 20 | V>A | No | gnomAD | |
| rs1815451168 | 21 | G>R | No | TOPMed | |
| rs1222321552 | 22 | V>L | No | gnomAD | |
| rs1222321552 | 22 | V>M | No | gnomAD | |
| rs1419211674 | 23 | F>L | No | Ensembl | |
| rs1279739765 | 23 | F>S | No |
TOPMed gnomAD |
|
| rs373638266 | 24 | Q>* | No | gnomAD | |
| rs1463437168 | 24 | Q>P | No |
TOPMed gnomAD |
|
| rs1815449275 | 25 | H>D | No | TOPMed | |
| rs369425515 | 25 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3861838 rs762027845 |
25 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs763355013 | 26 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1357313252 | 27 | K>N | No | gnomAD | |
| rs1323695286 | 28 | V>G | No | gnomAD | |
| rs1461206739 | 30 | I>T | No | TOPMed | |
| rs1298381462 | 31 | I>V | No | TOPMed | |
| rs745365008 | 32 | A>P | No |
ExAC TOPMed gnomAD |
|
|
rs745365008 COSM1443376 |
32 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs201814236 | 33 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1183924388 | 34 | D>E | No | gnomAD | |
| rs746628579 | 34 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs746628579 | 34 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs777342418 | 35 | Q>* | No |
ExAC gnomAD |
|
| rs1815446097 | 35 | Q>R | No | TOPMed | |
| rs1255972636 | 36 | G>A | No |
TOPMed gnomAD |
|
| rs1255972636 | 36 | G>D | No |
TOPMed gnomAD |
|
| rs1815445696 | 37 | N>T | No | TOPMed | |
| rs530017660 | 38 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs530017660 | 38 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs373639301 | 38 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs150810052 | 39 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs150810052 | 39 | T>P | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM483875 rs753725330 |
40 | T>I | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs753725330 | 40 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs753725330 | 40 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs767593530 | 41 | P>S | No |
ExAC gnomAD |
|
| rs1562343047 | 42 | S>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757305369 | 43 | Y>F | No |
ExAC gnomAD |
|
| rs189710538 | 44 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs762535261 | 45 | A>T | No | Ensembl | |
| rs763297872 | 47 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs763297872 | 47 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1000603059 | 50 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs903617843 | 51 | R>Q | No | TOPMed | |
|
rs760051391 COSM3346986 |
51 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1172905951 | 52 | L>P | No |
TOPMed gnomAD |
|
| rs141349016 | 53 | I>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1582677 rs141349016 |
53 | I>T | stomach [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs1179879737 | 54 | G>R | No | gnomAD | |
| rs1459435022 | 54 | G>V | No | gnomAD | |
| TCGA novel | 55 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757351933 | 56 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs771910474 | 57 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs772996658 | 57 | A>S | No |
ExAC gnomAD |
|
| TCGA novel | 58 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772862216 | 59 | N>K | No | gnomAD | |
| rs1815440069 | 61 | V>I | No | TOPMed | |
| rs1314037039 | 62 | A>T | No | gnomAD | |
| rs754849874 | 63 | M>I | No |
ExAC gnomAD |
|
| rs778784868 | 63 | M>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 64 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749200537 | 64 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1815438987 | 65 | P>H | No | Ensembl | |
|
rs1815438987 TCGA novel |
65 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1456569884 | 66 | Q>H | No | gnomAD | |
| rs1295575618 | 66 | Q>R | No | gnomAD | |
| rs757327606 | 67 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs1159555371 | 67 | N>S | No | gnomAD | |
| rs751654845 | 68 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs764284547 | 69 | V>G | No |
ExAC gnomAD |
|
| TCGA novel | 70 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1815437133 | 71 | D>G | No | Ensembl | |
| rs1815436574 | 73 | K>N | No | Ensembl | |
| rs1211282132 | 73 | K>Q | No | gnomAD | |
| rs145946517 | 74 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs371434396 | 74 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs962754902 | 76 | I>F | No | Ensembl | |
| rs1581697066 | 78 | R>S | No | Ensembl | |
| rs759870563 | 79 | K>E | No |
ExAC gnomAD |
|
| TCGA novel | 79 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM281785 | 79 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1213659188 | 81 | N>H | No |
TOPMed gnomAD |
|
| rs1355558679 | 83 | P>L | No | gnomAD | |
| rs1307041606 | 84 | V>A | No |
TOPMed gnomAD |
|
| rs777167633 | 85 | V>E | No |
ExAC TOPMed gnomAD |
|
| rs1461322233 | 85 | V>I | No | gnomAD | |
| rs766866292 | 86 | Q>* | No |
ExAC gnomAD |
|
| rs766866292 | 86 | Q>K | No |
ExAC gnomAD |
|
| rs772792188 | 87 | A>E | No |
ExAC gnomAD |
|
| rs760220858 | 87 | A>S | No |
ExAC gnomAD |
|
| rs1452233259 | 88 | D>V | No |
TOPMed gnomAD |
|
| rs1365145150 | 91 | L>F | No | gnomAD | |
| rs199562102 | 93 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1815429822 | 93 | P>S | No | gnomAD | |
| rs1359025519 | 94 | F>S | No | gnomAD | |
| rs1158559661 | 97 | I>V | No | gnomAD | |
| COSM1546241 | 99 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815428490 | 100 | G>E | No | Ensembl | |
| rs774168279 | 100 | G>R | No |
ExAC gnomAD |
|
| rs768475218 | 101 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs749076420 | 102 | K>M | No |
ExAC gnomAD |
|
| rs749076420 | 102 | K>T | No |
ExAC gnomAD |
|
| rs375770555 | 103 | P>S | No | Ensembl | |
| TCGA novel | 106 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1815427757 | 106 | L>P | No | TOPMed | |
| rs985952759 | 107 | V>G | No |
TOPMed gnomAD |
|
| rs1815427576 | 107 | V>M | No | TOPMed | |
| TCGA novel | 108 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769675691 | 109 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1022132419 | 109 | Y>C | No | TOPMed | |
| rs368011012 | 110 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1230308618 | 111 | G>R | No | gnomAD | |
|
COSM4391239 rs1815425901 |
112 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1815425714 | 113 | N>S | No | Ensembl | |
| rs1344564634 | 115 | A>V | No | TOPMed | |
| rs1345541682 | 117 | Y>C | No |
TOPMed gnomAD |
|
| rs758498452 | 117 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1339887302 | 118 | P>L | No | gnomAD | |
| COSM4923860 | 119 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM394318 rs1030228955 |
123 | S>L | lung Variant assessed as Somatic; MODERATE impact. endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs142437638 | 124 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs755368492 | 127 | T>S | No |
ExAC gnomAD |
|
| rs1402066769 | 129 | L>M | No |
TOPMed gnomAD |
|
| rs1402066769 | 129 | L>V | No |
TOPMed gnomAD |
|
| rs1815422724 | 131 | E>D | No | Ensembl | |
| rs1562342507 | 132 | T>I | No | Ensembl | |
| rs1222171982 | 133 | A>T | No |
TOPMed gnomAD |
|
| rs754215438 | 135 | A>D | No |
ExAC gnomAD |
|
| rs1815421772 | 136 | F>S | No | gnomAD | |
| rs1815421033 | 139 | H>Y | No | TOPMed | |
| rs1404919179 | 140 | P>A | No | gnomAD | |
| rs1815420090 | 142 | T>I | No | gnomAD | |
| rs761456778 | 144 | A>V | No | ExAC | |
| rs1194003547 | 146 | I>F | No | gnomAD | |
| rs747651249 | 147 | T>I | No | TOPMed | |
| rs1815418138 | 148 | V>A | No | gnomAD | |
| rs768259019 | 148 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM3861834 | 149 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs762791790 | 150 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs201394277 | 150 | A>V | No | 1000Genomes | |
| rs1280708557 | 153 | N>K | No |
TOPMed gnomAD |
|
| rs775085801 | 153 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs765590853 | 154 | D>E | No |
ExAC gnomAD |
|
| rs1046010671 | 156 | Q>L | No | TOPMed | |
| rs202165632 | 157 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs772111227 | 157 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs202165632 | 157 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1337651485 | 158 | Q>R | No |
TOPMed gnomAD |
|
| rs1815415131 | 159 | A>P | No | TOPMed | |
| rs1161195438 | 161 | K>E | No | TOPMed | |
| rs779186102 | 161 | K>N | No |
ExAC gnomAD |
|
| rs564894041 | 162 | D>Y | No |
ExAC gnomAD |
|
| rs1392095004 | 163 | A>E | No | TOPMed | |
| TCGA novel | 163 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1392095004 | 163 | A>V | No | TOPMed | |
| rs916118865 | 164 | G>D | No |
TOPMed gnomAD |
|
| rs764022454 | 165 | V>G | No | gnomAD | |
| rs754094690 | 165 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs780242965 | 166 | I>T | No |
ExAC gnomAD |
|
| rs145104527 | 167 | A>D | No |
ESP TOPMed |
|
| rs145104527 | 167 | A>V | No |
ESP TOPMed |
|
| rs1815412147 | 168 | G>E | No |
TOPMed gnomAD |
|
| rs149204208 | 170 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1815411883 | 170 | N>S | No |
TOPMed gnomAD |
|
| rs1815411883 | 170 | N>T | No |
TOPMed gnomAD |
|
| rs1473025198 | 172 | L>R | No | gnomAD | |
| rs1440265955 | 174 | I>S | No | TOPMed | |
| rs761403757 | 176 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs768176285 | 176 | N>Y | No |
ExAC gnomAD |
|
| rs777178387 | 177 | E>K | No | TOPMed | |
| rs777178387 | 177 | E>Q | No | TOPMed | |
| COSM1329799 | 178 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815408864 | 178 | P>S | No | TOPMed | |
| rs138274398 | 179 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1313297132 | 180 | A>G | No |
TOPMed gnomAD |
|
| rs1179576532 | 183 | I>T | No | gnomAD | |
| rs769126652 | 183 | I>V | No | TOPMed | |
| rs764922859 | 185 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs571006499 | 185 | Y>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1815406315 | 187 | L>V | No | Ensembl | |
| rs1448410696 | 188 | D>H | No | gnomAD | |
| rs747436894 | 189 | K>R | No |
TOPMed gnomAD |
|
|
rs776550285 COSM3702978 |
190 | G>A | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| TCGA novel | 192 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1405533450 | 193 | G>E | No | gnomAD | |
| COSM3624803 | 193 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 194 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 194 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770917469 | 195 | R>* | No |
ExAC TOPMed gnomAD |
|
|
rs201198988 RCV000970452 |
195 | R>Q | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs768814318 | 196 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs774228634 | 196 | H>R | No |
ExAC gnomAD |
|
| rs1180401356 | 199 | I>V | No | gnomAD | |
| rs373875697 | 201 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1815403798 | 202 | L>V | No | TOPMed | |
| rs1815402944 | 204 | G>E | No | Ensembl | |
| rs1232357390 | 204 | G>R | No |
TOPMed gnomAD |
|
| rs1165445231 | 205 | G>D | No |
TOPMed gnomAD |
|
| rs780372000 | 206 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1581696153 | 206 | T>I | No | Ensembl | |
| rs756514211 | 207 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1290081406 | 208 | D>N | No | gnomAD | |
| rs1222147845 | 208 | D>V | No | gnomAD | |
| rs757776898 | 209 | V>G | No |
ExAC gnomAD |
|
| rs781761072 | 209 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs781761072 | 209 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1370561662 | 211 | I>T | No | gnomAD | |
| rs1815400485 | 211 | I>V | No | Ensembl | |
| rs752112742 | 212 | L>P | No |
ExAC gnomAD |
|
| rs1815399309 | 213 | T>I | No | Ensembl | |
| rs1815399515 | 213 | T>P | No | TOPMed | |
| rs762890249 | 214 | I>T | No |
TOPMed gnomAD |
|
|
COSM3624802 rs1581696037 |
215 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1815397862 | 216 | D>G | No | TOPMed | |
| TCGA novel | 219 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1300880929 | 220 | E>* | No | gnomAD | |
| rs150472288 | 221 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752264409 | 224 | T>S | No |
ExAC gnomAD |
|
| rs2151461615 | 227 | D>Y | No | 1000Genomes | |
| rs548569388 | 229 | H>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1815394487 | 229 | H>Q | No | Ensembl | |
| rs759317010 | 230 | L>P | No |
ExAC gnomAD |
|
| rs759317010 | 230 | L>R | No |
ExAC gnomAD |
|
| rs371447094 | 231 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs773226597 | 235 | F>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM6173933 | 236 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768598138 | 237 | N>S | No |
ExAC gnomAD |
|
| rs2151461569 | 238 | R>S | No | Ensembl | |
| rs191859553 | 238 | R>T | No | 1000Genomes | |
| COSM1443369 | 239 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2151461558 | 240 | V>A | No | 1000Genomes | |
| rs1281058142 | 240 | V>L | No | gnomAD | |
|
rs1403940171 COSM1294289 |
241 | S>N | cervix [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs775568758 | 241 | S>R | No |
ExAC gnomAD |
|
| rs1234410625 | 242 | H>L | No |
TOPMed gnomAD |
|
| rs1355403980 | 242 | H>Y | No |
TOPMed gnomAD |
|
| rs1290344128 | 243 | F>V | No | gnomAD | |
| rs147868862 | 244 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs147868862 | 244 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs781631922 | 245 | E>K | No |
ExAC gnomAD |
|
| COSM71107 | 246 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs911295867 | 248 | K>Q | No | Ensembl | |
|
COSM3921399 rs1271868470 |
249 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs747422784 | 251 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1815389384 | 252 | K>Q | No | gnomAD | |
| TCGA novel | 253 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1815388792 | 254 | D>N | No |
TOPMed gnomAD |
|
| rs1815388422 | 255 | I>M | No | TOPMed | |
| COSM4911785 | 255 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815388243 | 256 | S>G | No | TOPMed | |
| rs200543237 | 257 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs757844713 | 259 | K>R | No |
ExAC gnomAD |
|
| rs764743827 | 260 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs764743827 | 260 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs199646393 | 260 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs199646393 | 260 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 262 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM185487 rs142416335 |
262 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM1568415 | 263 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750295996 | 264 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs760512895 | 264 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs199532053 | 266 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs775580678 | 266 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs775580678 | 266 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs769994484 | 267 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs142613640 | 270 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs148442705 | 272 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1184127823 | 273 | K>E | No | gnomAD | |
| rs1408091890 | 276 | L>P | No | gnomAD | |
|
COSM3624799 rs1460318497 |
277 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs748610630 | 278 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs986208345 | 279 | S>G | No | TOPMed | |
| COSM2157007 | 281 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815381952 | 282 | A>G | No | Ensembl | |
| rs754506860 | 283 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs1815381417 | 283 | N>K | No | Ensembl | |
|
rs1581695526 COSM387641 |
283 | N>S | lung [Cosmic] | No |
cosmic curated Ensembl |
| rs952995140 | 285 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1306759439 | 286 | I>T | No |
TOPMed gnomAD |
|
| rs1250671657 | 286 | I>V | No |
TOPMed gnomAD |
|
| rs779724006 | 287 | D>Y | No |
ExAC gnomAD |
|
| rs187074836 | 289 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs187074836 | 289 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767230108 | 292 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs989377237 | 292 | G>S | No |
TOPMed gnomAD |
|
| rs1489375367 | 293 | I>V | No |
TOPMed gnomAD |
|
|
VAR_025843 rs34360259 |
294 | D>G | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1296725819 | 295 | F>S | No | gnomAD | |
| rs1435433434 | 295 | F>V | No |
TOPMed gnomAD |
|
| rs1461620456 | 297 | T>I | No |
TOPMed gnomAD |
|
| rs1815377085 | 299 | I>F | No | Ensembl | |
| rs1382132987 | 301 | R>K | No | TOPMed | |
| rs1815376598 | 301 | R>S | No | Ensembl | |
| TCGA novel | 301 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765356075 | 302 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs765356075 | 302 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1815376213 | 303 | R>* | No | TOPMed | |
|
rs1385211232 COSM4940390 |
303 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1815375694 | 306 | E>D | No | Ensembl | |
| rs368299209 | 308 | C>Y | No | gnomAD | |
| rs1193728135 | 309 | A>T | No |
TOPMed gnomAD |
|
| rs1480067363 | 309 | A>V | No | gnomAD | |
| rs546676852 | 310 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs1815373824 | 312 | F>C | No | Ensembl | |
| rs1356972985 | 313 | R>K | No | gnomAD | |
| rs941324389 | 314 | G>C | No |
TOPMed gnomAD |
|
| rs1240769641 | 314 | G>D | No | gnomAD | |
| rs773562809 | 317 | E>K | No | ExAC | |
| rs748640312 | 319 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1815372685 | 320 | E>G | No | gnomAD | |
| rs1581695219 | 321 | K>N | No | Ensembl | |
| rs779447554 | 322 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM300531 rs768126334 |
322 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1365331743 | 323 | L>R | No | gnomAD | |
| rs142398035 | 324 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs779666881 COSM4514337 |
324 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1178988304 | 325 | D>G | No |
TOPMed gnomAD |
|
| rs750047577 | 325 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs750047577 | 325 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs780995765 | 326 | A>G | No |
ExAC TOPMed gnomAD |
|
| COSM3624797 | 328 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815370271 | 329 | D>G | No | TOPMed | |
| rs976294956 | 329 | D>H | No | gnomAD | |
| rs976294956 | 329 | D>N | No | gnomAD | |
| rs757021527 | 330 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1369822811 | 331 | A>G | No |
TOPMed gnomAD |
|
| rs1369822811 | 331 | A>V | No |
TOPMed gnomAD |
|
| rs1815369718 | 334 | H>N | No |
TOPMed gnomAD |
|
| rs1290691195 | 335 | D>G | No | gnomAD | |
| rs772366553 | 335 | D>N | No |
TOPMed gnomAD |
|
| rs764164396 | 336 | I>T | No |
ExAC gnomAD |
|
| rs751517866 | 336 | I>V | No |
ExAC gnomAD |
|
| rs1259058990 | 338 | L>F | No |
TOPMed gnomAD |
|
| rs1302824566 | 339 | V>A | No |
TOPMed gnomAD |
|
| rs1314611285 | 341 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs755098112 | 341 | G>R | No |
ExAC gnomAD |
|
| rs755098112 | 341 | G>S | No |
ExAC gnomAD |
|
| rs1314611285 | 341 | G>V | No |
TOPMed gnomAD |
|
| rs1815367947 | 342 | S>A | No | gnomAD | |
| rs1261254929 | 342 | S>F | No |
TOPMed gnomAD |
|
| rs1581694965 | 343 | T>I | No | Ensembl | |
| rs1581694965 | 343 | T>N | No | Ensembl | |
| rs544557163 | 344 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs544557163 | 344 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs375290879 COSM4752165 |
344 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP NCI-TCGA TOPMed gnomAD |
| rs375290879 | 344 | R>P | No |
ESP TOPMed gnomAD |
|
| rs1345287525 | 345 | I>V | No | gnomAD | |
| rs1384450016 | 346 | P>L | No | gnomAD | |
| rs760914999 | 347 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs760914999 | 347 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs547752581 | 349 | Q>E | No | Ensembl | |
| rs1815365518 | 349 | Q>R | No | Ensembl | |
| rs923849656 | 350 | R>Q | No | gnomAD | |
| rs1429370855 | 350 | R>W | No |
TOPMed gnomAD |
|
| rs773329064 | 351 | L>P | No |
ExAC gnomAD |
|
| rs762126324 | 352 | L>V | No |
ExAC gnomAD |
|
| rs1259619117 | 354 | D>G | No | gnomAD | |
| rs1815363921 | 356 | F>L | No | Ensembl | |
| rs181670945 | 357 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs529670905 | 359 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1036938945 | 359 | R>H | No | gnomAD | |
| rs1815362432 | 360 | D>V | No | TOPMed | |
| rs967686617 | 362 | N>K | No |
TOPMed gnomAD |
|
| rs944858395 | 363 | K>Q | No |
TOPMed gnomAD |
|
| rs911644560 | 363 | K>R | No |
TOPMed gnomAD |
|
| rs1460288819 | 364 | S>N | No |
TOPMed gnomAD |
|
| rs1406184281 | 367 | P>A | No |
TOPMed gnomAD |
|
| rs1338085795 | 368 | D>E | No |
TOPMed gnomAD |
|
| rs137935258 | 368 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1428221917 | 370 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1815359486 | 374 | G>R | No | Ensembl | |
| TCGA novel | 375 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1461660496 | 376 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1815358424 | 378 | Q>* | No | TOPMed | |
| rs757043144 | 378 | Q>H | No |
ExAC gnomAD |
|
| rs746861272 | 380 | A>P | No |
ExAC gnomAD |
|
| rs1216535612 | 381 | I>T | No | gnomAD | |
| rs1486169523 | 383 | M>T | No |
TOPMed gnomAD |
|
| rs1269911192 | 383 | M>V | No | gnomAD | |
| rs146199357 | 384 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1815356457 | 386 | K>E | No |
TOPMed gnomAD |
|
| rs141209981 | 388 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1449880562 | 388 | E>D | No | gnomAD | |
|
COSM1294273 rs766434444 |
389 | K>N | cervix [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs756198272 | 390 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs756198272 | 390 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs1292518532 COSM3413928 |
391 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1292518532 | 391 | Q>E | No |
TOPMed gnomAD |
|
| rs1815354632 | 396 | L>R | No | TOPMed | |
| rs1815354262 | 397 | D>G | No |
TOPMed gnomAD |
|
| rs139384991 | 398 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139384991 | 398 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1480405141 | 399 | A>T | No | Ensembl | |
| rs555125949 | 399 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs1267735516 | 400 | P>L | No | gnomAD | |
| rs1815353072 | 400 | P>S | No | TOPMed | |
| rs1815353072 | 400 | P>T | No | TOPMed | |
| rs1234258051 | 402 | S>C | No | gnomAD | |
| TCGA novel | 402 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372714721 | 404 | G>E | No |
ESP TOPMed gnomAD |
|
| rs566332175 | 407 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776340641 | 408 | A>T | No |
ExAC TOPMed gnomAD |
|
|
COSM4929396 rs1815350523 |
409 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs770435641 | 410 | G>D | No |
ExAC gnomAD |
|
| rs1245863225 | 410 | G>S | No | gnomAD | |
| rs777675880 | 411 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs758295721 | 412 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs1412918753 | 412 | M>V | No | TOPMed | |
| rs17857014 | 416 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs778757324 | 417 | K>* | No |
ExAC gnomAD |
|
| rs756216078 | 418 | R>C | No |
ExAC gnomAD |
|
|
COSM3783977 rs1815348063 |
418 | R>H | Variant assessed as Somatic; MODERATE impact. prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1161610466 | 419 | N>S | No | gnomAD | |
| rs1161610466 | 419 | N>T | No | gnomAD | |
| rs2151460728 | 421 | T>I | No | Ensembl | |
| rs1408852268 | 423 | P>S | No | gnomAD | |
| rs1815347135 | 424 | T>A | No | Ensembl | |
| COSM3928341 | 424 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1178976039 | 425 | K>R | No | gnomAD | |
| rs1387287493 | 428 | Q>E | No | gnomAD | |
| rs1815345987 | 430 | F>S | No | Ensembl | |
| rs367836600 | 431 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs367836600 | 431 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1222406248 | 432 | T>A | No | gnomAD | |
| rs1815345131 | 433 | Y>F | No | gnomAD | |
| rs757403494 | 435 | D>E | No |
ExAC gnomAD |
|
| rs1172737962 | 435 | D>V | No | gnomAD | |
| rs1307180997 | 437 | Q>* | No | TOPMed | |
| rs1307180997 | 437 | Q>K | No | TOPMed | |
| rs751809556 | 438 | P>R | No |
ExAC gnomAD |
|
| rs571792102 | 439 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM6039506 | 440 | V>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201161706 | 440 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs201161706 | 440 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1345768587 | 441 | L>P | No | gnomAD | |
| rs1161166376 | 442 | I>N | No | gnomAD | |
| TCGA novel | 443 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1177431221 | 444 | V>M | No | gnomAD | |
| COSM4927775 | 445 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4691344 rs770600264 |
448 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs760358503 | 449 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1815340853 | 450 | A>T | No | TOPMed | |
| rs1244372817 | 451 | M>T | No |
TOPMed gnomAD |
|
| rs1815340154 | 455 | N>D | No | TOPMed | |
| rs773048128 | 455 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs773048128 | 455 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs773048128 | 455 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs771963411 | 458 | L>P | No |
ExAC gnomAD |
|
| rs898088646 | 459 | G>E | No |
TOPMed gnomAD |
|
| rs748024336 | 459 | G>R | No |
ExAC gnomAD |
|
| rs147667814 | 460 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs147667814 | 460 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1815338743 | 460 | R>W | No | TOPMed | |
| rs1815338209 | 462 | D>E | No | TOPMed | |
|
rs1432329964 COSM1294259 |
464 | T>S | cervix [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
TCGA novel rs1815337716 |
465 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1815337547 | 467 | P>R | No | TOPMed | |
| rs1387998646 | 470 | P>S | No | gnomAD | |
| rs1391933409 | 471 | R>G | No |
TOPMed gnomAD |
|
| rs749251656 | 472 | G>E | No |
ExAC gnomAD |
|
| rs781091208 | 473 | V>F | No |
ExAC gnomAD |
|
| rs778008112 | 477 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs778008112 | 477 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs758754760 | 477 | E>V | No |
ExAC gnomAD |
|
| rs1201212729 | 478 | V>A | No |
TOPMed gnomAD |
|
|
rs482145 VAR_025844 RCV000965703 |
479 | T>M | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1485804840 | 480 | F>L | No | gnomAD | |
| rs1815334415 | 481 | D>V | No | gnomAD | |
| rs754395053 | 482 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1300116883 | 483 | D>G | No | Ensembl | |
| TCGA novel | 483 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs548364201 | 484 | A>G | No | gnomAD | |
| rs548364201 | 484 | A>V | No | gnomAD | |
| rs1815333272 | 485 | N>H | No |
TOPMed gnomAD |
|
| rs765970026 | 485 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1815332675 | 488 | L>F | No | Ensembl | |
| rs1815332675 | 488 | L>I | No | Ensembl | |
| rs1212762938 | 488 | L>P | No | gnomAD | |
| rs773948561 | 489 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1343407673 | 491 | T>K | No | gnomAD | |
| rs1815331520 | 491 | T>S | No | TOPMed | |
| rs1356674722 | 492 | A>D | No | gnomAD | |
| rs780016316 | 493 | T>A | No |
ExAC TOPMed gnomAD |
|
|
VAR_003820 rs2227956 |
493 | T>M | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2227956 | 493 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM1443364 | 494 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000929873 rs576268420 |
495 | K>missing | No |
ClinVar dbSNP |
|
| rs972516481 | 495 | K>E | No | Ensembl | |
| rs1815329333 | 496 | S>G | No | TOPMed | |
| rs2151460279 | 496 | S>T | No | Ensembl | |
| rs758555654 | 498 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1240661889 | 498 | G>D | No | gnomAD | |
| rs758555654 | 498 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1240661889 | 498 | G>V | No | gnomAD | |
| rs2151460244 | 499 | K>Q | No | Ensembl | |
| rs748435926 | 499 | K>R | No |
ExAC gnomAD |
|
| rs1202244137 | 500 | V>A | No | gnomAD | |
| rs1202244137 | 500 | V>E | No | gnomAD | |
| rs1252877703 | 500 | V>M | No | gnomAD | |
| rs1815327696 | 503 | I>S | No | Ensembl | |
| rs755419733 | 504 | T>A | No |
ExAC gnomAD |
|
| rs1815326932 | 505 | I>M | No | TOPMed | |
| rs200301187 | 505 | I>V | No |
1000Genomes gnomAD |
|
| rs17853300 | 506 | T>A | No | Ensembl | |
| rs934950628 | 506 | T>N | No | TOPMed | |
| rs754272971 | 507 | N>D | No |
ExAC gnomAD |
|
| rs767063723 | 508 | D>H | No |
ExAC gnomAD |
|
| rs1562339863 | 508 | D>V | No | Ensembl | |
| rs1375867687 | 509 | K>E | No | gnomAD | |
| rs755676343 | 509 | K>N | No |
ExAC TOPMed gnomAD |
|
| COSM3624796 | 510 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs148694457 | 511 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767137456 | 511 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs148694457 | 511 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1166029147 | 513 | S>T | No | gnomAD | |
| rs1028585381 | 514 | K>N | No | gnomAD | |
| rs998578135 | 515 | E>D | No | gnomAD | |
| rs761329208 | 516 | E>D | No |
ExAC gnomAD |
|
| COSM3346939 | 516 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs111279147 | 517 | I>M | No |
1000Genomes ExAC gnomAD |
|
| rs774072389 | 517 | I>T | No |
ExAC gnomAD |
|
| rs139969031 | 519 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM5870208 rs775401949 |
519 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs769779342 | 520 | M>V | No |
ExAC gnomAD |
|
| rs745816477 | 521 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1815322318 | 522 | L>M | No | TOPMed | |
| rs1581693313 | 522 | L>Q | No | Ensembl | |
| rs1581693306 | 523 | D>E | No | Ensembl | |
| rs1815321850 | 524 | A>S | No | Ensembl | |
| rs544006150 | 524 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs1815321328 | 525 | E>K | No | Ensembl | |
| rs1305206605 | 527 | Y>C | No |
TOPMed gnomAD |
|
| rs1318813778 | 527 | Y>H | No |
TOPMed gnomAD |
|
| rs1815320500 | 528 | K>E | No |
TOPMed gnomAD |
|
| rs1409382230 | 528 | K>R | No | gnomAD | |
| rs1409382230 | 528 | K>T | No | gnomAD | |
| rs779235392 | 529 | A>G | No |
ExAC gnomAD |
|
| rs755297837 | 530 | E>K | No |
ExAC gnomAD |
|
| COSM4830030 | 531 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1466886931 | 532 | E>D | No |
TOPMed gnomAD |
|
| COSM4529208 | 532 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815319551 | 533 | V>F | No |
TOPMed gnomAD |
|
| rs1815319261 | 534 | Q>H | No | Ensembl | |
| rs1403657550 | 534 | Q>R | No | gnomAD | |
| COSM741851 | 535 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201132627 | 536 | E>K | No |
1000Genomes gnomAD |
|
| rs201132627 | 536 | E>Q | No |
1000Genomes gnomAD |
|
| rs1246847723 | 538 | I>M | No |
TOPMed gnomAD |
|
| rs982029613 | 538 | I>T | No |
TOPMed gnomAD |
|
| rs998930397 | 540 | A>S | No | Ensembl | |
| rs554869938 | 540 | A>V | No |
1000Genomes ExAC gnomAD |
|
| COSM1077705 | 541 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780581828 | 544 | L>* | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 545 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756624684 | 546 | S>C | No |
ExAC gnomAD |
|
| rs756624684 | 546 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs146531367 | 547 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs146531367 | 547 | Y>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1156514355 | 547 | Y>H | No | gnomAD | |
|
rs146402090 COSM238588 |
548 | A>D | prostate [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs146402090 | 548 | A>G | No |
TOPMed gnomAD |
|
| rs1429767841 | 548 | A>P | No | gnomAD | |
|
rs146402090 COSM109759 |
548 | A>V | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs111462391 | 550 | N>S | No | gnomAD | |
| rs1391977656 | 551 | M>I | No |
TOPMed gnomAD |
|
| rs1161163512 | 551 | M>T | No | TOPMed | |
| TCGA novel | 552 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1815315620 | 557 | D>E | No | Ensembl | |
| TCGA novel | 557 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs566393477 | 558 | E>D | No |
1000Genomes ExAC gnomAD |
|
| rs756836668 | 558 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs762641185 | 559 | G>D | No |
ExAC gnomAD |
|
| rs1324473563 | 560 | L>M | No |
TOPMed gnomAD |
|
| rs1815314165 | 563 | K>N | No | Ensembl | |
| rs765019123 | 563 | K>R | No |
ExAC gnomAD |
|
| rs1815313739 | 566 | E>D | No | TOPMed | |
| rs199974706 | 566 | E>G | No | Ensembl | |
| rs1351762990 | 567 | S>F | No | gnomAD | |
|
rs567637853 RCV000895789 |
568 | D>* | No |
ClinVar dbSNP |
|
| rs1815313426 | 569 | K>* | No | Ensembl | |
| rs1440150593 | 569 | K>T | No | Ensembl | |
| rs939012192 | 573 | L>S | No |
TOPMed gnomAD |
|
| TCGA novel | 573 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs939012192 | 573 | L>W | No |
TOPMed gnomAD |
|
| rs759422957 | 574 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1230738576 | 576 | C>F | No | TOPMed | |
| rs776599600 | 576 | C>G | No |
ExAC gnomAD |
|
| rs1815311550 | 577 | N>D | No | Ensembl | |
| rs748331299 | 577 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs772013963 | 577 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs772013963 | 577 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs554479408 | 578 | E>D | No |
1000Genomes ExAC gnomAD |
|
| rs139052456 | 578 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1487613 | 578 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815310364 | 580 | L>V | No | TOPMed | |
| rs1815310214 | 581 | S>A | No | Ensembl | |
|
rs371717441 COSM1443362 |
581 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1208238536 | 582 | W>* | No | gnomAD | |
| rs1815309511 | 582 | W>* | No | TOPMed | |
| rs1208238536 | 582 | W>C | No | gnomAD | |
| rs1246285394 | 582 | W>G | No | gnomAD | |
| TCGA novel | 582 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781740332 | 584 | E>G | No |
ExAC gnomAD |
|
| rs756717103 | 585 | V>G | No |
ExAC gnomAD |
|
| rs144451142 | 586 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1184253837 | 587 | Q>H | No |
TOPMed gnomAD |
|
| rs1815308532 | 587 | Q>K | No | TOPMed | |
| rs1271096786 | 587 | Q>P | No | gnomAD | |
| rs1271096786 | 587 | Q>R | No | gnomAD | |
| rs1230290569 | 590 | E>K | No | gnomAD | |
| rs777160535 | 592 | D>A | No |
ExAC gnomAD |
|
| rs758000575 | 592 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs202033031 | 592 | D>Y | No |
TOPMed gnomAD |
|
| rs61751215 | 593 | E>D | No |
ESP ExAC gnomAD |
|
| rs1338315015 | 594 | F>C | No | gnomAD | |
| rs1338315015 | 594 | F>S | No | gnomAD | |
| rs1815306532 | 595 | D>H | No | Ensembl | |
| COSM1312019 | 595 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 597 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1285348321 | 597 | K>N | No | gnomAD | |
| rs1815306223 | 597 | K>Q | No | Ensembl | |
| rs1412339351 | 597 | K>R | No | gnomAD | |
| rs1408402334 | 598 | R>* | No |
TOPMed gnomAD |
|
| rs201332987 | 598 | R>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201332987 | 598 | R>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1815304909 | 599 | K>N | No | TOPMed | |
| rs1338256513 | 599 | K>R | No |
TOPMed gnomAD |
|
| COSM6106560 | 602 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs2075800 VAR_025846 |
602 | E>K | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs954264438 | 604 | M>I | No |
TOPMed gnomAD |
|
| rs1388289260 | 604 | M>V | No | gnomAD | |
| rs371226757 | 605 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs371226757 | 605 | C>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs371226757 | 605 | C>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs774425727 | 606 | N>K | No |
ExAC gnomAD |
|
| COSM6106561 | 607 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1311612034 | 608 | I>V | No |
TOPMed gnomAD |
|
| rs1815303471 | 609 | I>V | No | Ensembl | |
| rs768651596 | 611 | K>Q | No |
ExAC gnomAD |
|
| rs1009138682 | 613 | Y>* | No |
TOPMed gnomAD |
|
| rs1581692670 | 613 | Y>H | No | Ensembl | |
| COSM4405560 | 614 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377450492 | 615 | G>R | No |
ESP ExAC gnomAD |
|
| rs1815302492 | 617 | C>R | No | TOPMed | |
| COSM6173934 | 619 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3624794 | 620 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1212406436 | 620 | P>L | No | Ensembl | |
| rs1358090171 | 620 | P>S | No | TOPMed | |
| COSM269333 | 621 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1815301119 | 623 | G>A | No |
TOPMed gnomAD |
|
|
rs746297288 COSM294455 |
623 | G>R | lung large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs771411091 | 625 | G>A | No | ExAC | |
| rs771411091 | 625 | G>V | No | ExAC | |
| rs777296245 | 626 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs777296245 | 626 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs1815299623 | 627 | V>A | No | Ensembl | |
| rs2151459665 | 628 | P>R | No | Ensembl | |
| rs1411517785 | 630 | R>K | No |
TOPMed gnomAD |
|
| rs757954206 | 630 | R>S | No |
ExAC gnomAD |
|
| rs202159508 | 631 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1387186082 | 632 | A>D | No |
TOPMed gnomAD |
|
| rs1387186082 | 632 | A>V | No |
TOPMed gnomAD |
|
| rs1431714253 | 633 | T>A | No |
TOPMed gnomAD |
|
| rs1815297915 | 633 | T>I | No |
TOPMed gnomAD |
|
| rs1021677888 | 634 | G>S | No | Ensembl | |
| rs1815297439 | 635 | P>H | No |
TOPMed gnomAD |
|
| TCGA novel | 636 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2151459627 | 637 | I>T | No | Ensembl | |
| rs370564561 | 637 | I>V | No |
ESP TOPMed gnomAD |
|
| rs753615095 | 639 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs753615095 | 639 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1482287694 | 640 | V>A | No |
TOPMed gnomAD |
|
| rs1482287694 | 640 | V>E | No |
TOPMed gnomAD |
|
| rs150873641 | 640 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1397100927 | 641 | D>G | No |
TOPMed gnomAD |
No associated diseases with P34931
3 regional properties for P34931
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Heat shock protein 70, conserved site | 11 - 18 | IPR018181-1 |
| conserved_site | Heat shock protein 70, conserved site | 199 - 212 | IPR018181-2 |
| conserved_site | Heat shock protein 70, conserved site | 336 - 350 | IPR018181-3 |
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| blood microparticle | A phospholipid microvesicle that is derived from any of several cell types, such as platelets, blood cells, endothelial cells, or others, and contains membrane receptors as well as other proteins characteristic of the parental cell. Microparticles are heterogeneous in size, and are characterized as microvesicles free of nucleic acids. |
| cell body | The portion of a cell bearing surface projections such as axons, dendrites, cilia, or flagella that includes the nucleus, but excludes all cell projections. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| mitochondrial matrix | The gel-like material, with considerable fine structure, that lies in the matrix space, or lumen, of a mitochondrion. It contains the enzymes of the tricarboxylic acid cycle and, in some organisms, the enzymes concerned with fatty acid oxidation. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| vesicle | Any small, fluid-filled, spherical organelle enclosed by membrane. |
| zona pellucida receptor complex | A multisubunit complex comprising the chaperonin-containing T-complex and several other components involved in mediating sperm-oocyte Interaction. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| ATP-dependent protein folding chaperone | Binding to a protein or a protein-containing complex to assist the protein folding process, driven by ATP hydrolysis. |
| heat shock protein binding | Binding to a heat shock protein, a protein synthesized or activated in response to heat shock. |
| protein folding chaperone | Binding to a protein or a protein-containing complex to assist the protein folding process. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
| unfolded protein binding | Binding to an unfolded protein. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| binding of sperm to zona pellucida | The process in which the sperm binds to the zona pellucida glycoprotein layer of the egg. The process begins with the attachment of the sperm plasma membrane to the zona pellucida and includes attachment of the acrosome inner membrane to the zona pellucida after the acrosomal reaction takes place. |
| chaperone cofactor-dependent protein refolding | The process of assisting in the correct posttranslational noncovalent assembly of proteins, which is dependent on additional protein cofactors. This process occurs over one or several cycles of nucleotide hydrolysis-dependent binding and release. |
| positive regulation of protein targeting to mitochondrion | Any process that activates or increases the frequency, rate or extent of protein targeting to mitochondrion. |
| protein refolding | The process carried out by a cell that restores the biological activity of an unfolded or misfolded protein, using helper proteins such as chaperones. |
| response to unfolded protein | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an unfolded protein stimulus. |
57 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P19120 | HSPA8 | Heat shock cognate 71 kDa protein | Bos taurus (Bovine) | SS |
| Q27975 | HSPA1A | Heat shock 70 kDa protein 1A | Bos taurus (Bovine) | SS |
| Q2YDD0 | HSPA14 | Heat shock 70 kDa protein 14 | Bos taurus (Bovine) | SS |
| P0CB32 | HSPA1L | Heat shock 70 kDa protein 1-like | Bos taurus (Bovine) | SS |
| O73885 | HSPA8 | Heat shock cognate 71 kDa protein | Gallus gallus (Chicken) | SS |
| E1C2P3 | HSPA14 | Heat shock 70 kDa protein 14 | Gallus gallus (Chicken) | SS |
| P08106 | Heat shock 70 kDa protein | Gallus gallus (Chicken) | SS | |
| P77319 | hscC | Chaperone protein HscC | Escherichia coli (strain K12) | PR |
| P0A6Y8 | dnaK | Chaperone protein DnaK | Escherichia coli (strain K12) | SS |
| P29843 | Hsc70-1 | Heat shock 70 kDa protein cognate 1 | Drosophila melanogaster (Fruit fly) | SS |
| P02825 | Hsp70Ab | Major heat shock 70 kDa protein Ab | Drosophila melanogaster (Fruit fly) | SS |
| P82910 | Hsp70Aa | Major heat shock 70 kDa protein Aa | Drosophila melanogaster (Fruit fly) | SS |
| Q8INI8 | Hsp70Ba | Major heat shock 70 kDa protein Ba | Drosophila melanogaster (Fruit fly) | SS |
| Q9BIR7 | Hsp70Bc | Major heat shock 70 kDa protein Bc | Drosophila melanogaster (Fruit fly) | SS |
| Q9BIS2 | Hsp70Bb | Major heat shock 70 kDa protein Bb | Drosophila melanogaster (Fruit fly) | SS |
| O97125 | Hsp68 | Heat shock protein 68 | Drosophila melanogaster (Fruit fly) | SS |
| P11146 | Hsc70-2 | Heat shock 70 kDa protein cognate 2 | Drosophila melanogaster (Fruit fly) | SS |
| P11147 | Hsc70-4 | Heat shock 70 kDa protein cognate 4 | Drosophila melanogaster (Fruit fly) | SS |
| Q9VG58 | Hsp70Bbb | Major heat shock 70 kDa protein Bbb | Drosophila melanogaster (Fruit fly) | SS |
| Q96MM6 | HSPA12B | Heat shock 70 kDa protein 12B | Homo sapiens (Human) | PR |
| O43301 | HSPA12A | Heat shock 70 kDa protein 12A | Homo sapiens (Human) | PR |
| P0DMV9 | HSPA1B | Heat shock 70 kDa protein 1B | Homo sapiens (Human) | SS |
| P0DMV8 | HSPA1A | Heat shock 70 kDa protein 1A | Homo sapiens (Human) | SS |
| P17066 | HSPA6 | Heat shock 70 kDa protein 6 | Homo sapiens (Human) | SS |
| P54652 | HSPA2 | Heat shock-related 70 kDa protein 2 | Homo sapiens (Human) | SS |
| Q0VDF9 | HSPA14 | Heat shock 70 kDa protein 14 | Homo sapiens (Human) | SS |
| P38646 | HSPA9 | Stress-70 protein, mitochondrial | Homo sapiens (Human) | SS |
| P11021 | HSPA5 | Endoplasmic reticulum chaperone BiP | Homo sapiens (Human) | SS |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | Homo sapiens (Human) | EV |
| P48741 | HSPA7 | Putative heat shock 70 kDa protein 7 | Homo sapiens (Human) | SS |
| P11143 | HSP70 | Heat shock 70 kDa protein | Zea mays (Maize) | SS |
| P63017 | Hspa8 | Heat shock cognate 71 kDa protein | Mus musculus (Mouse) | SS |
| Q99M31 | Hspa14 | Heat shock 70 kDa protein 14 | Mus musculus (Mouse) | SS |
| Q8K0U4 | Hspa12a | Heat shock 70 kDa protein 12A | Mus musculus (Mouse) | PR |
| P17156 | Hspa2 | Heat shock-related 70 kDa protein 2 | Mus musculus (Mouse) | SS |
| Q61696 | Hspa1a | Heat shock 70 kDa protein 1A | Mus musculus (Mouse) | SS |
| Q9CZJ2 | Hspa12b | Heat shock 70 kDa protein 12B | Mus musculus (Mouse) | PR |
| P17879 | Hspa1b | Heat shock 70 kDa protein 1B | Mus musculus (Mouse) | SS |
| P16627 | Hspa1l | Heat shock 70 kDa protein 1-like | Mus musculus (Mouse) | SS |
| Q6S4N2 | HSPA1B | Heat shock 70 kDa protein 1B | Sus scrofa (Pig) | SS |
| P14659 | Hspa2 | Heat shock-related 70 kDa protein 2 | Rattus norvegicus (Rat) | SS |
| P0DMW1 | Hspa1b | Heat shock 70 kDa protein 1B | Rattus norvegicus (Rat) | SS |
| P0DMW0 | Hspa1a | Heat shock 70 kDa protein 1A | Rattus norvegicus (Rat) | SS |
| P63018 | Hspa8 | Heat shock cognate 71 kDa protein | Rattus norvegicus (Rat) | SS |
| P55063 | Hspa1l | Heat shock 70 kDa protein 1-like | Rattus norvegicus (Rat) | SS |
| P09446 | hsp-1 | Heat shock protein hsp-1 | Caenorhabditis elegans | SS |
| P26413 | HSP70 | Heat shock 70 kDa protein | Glycine max (Soybean) (Glycine hispida) | SS |
| O65719 | HSP70-3 | Heat shock 70 kDa protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P22954 | HSP70-2 | Heat shock 70 kDa protein 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9C7X7 | HSP70-18 | Heat shock 70 kDa protein 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LHA8 | HSP70-4 | Heat shock 70 kDa protein 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9S9N1 | HSP70-5 | Heat shock 70 kDa protein 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P22953 | HSP70-1 | Heat shock 70 kDa protein 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P24629 | HSC-I | Heat shock cognate 70 kDa protein 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| P27322 | HSC-2 | Heat shock cognate 70 kDa protein 2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q5RGE6 | hspa14 | Heat shock 70 kDa protein 14 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90473 | hspa8 | Heat shock cognate 71 kDa protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MATAKGIAIG | IDLGTTYSCV | GVFQHGKVEI | IANDQGNRTT | PSYVAFTDTE | RLIGDAAKNQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VAMNPQNTVF | DAKRLIGRKF | NDPVVQADMK | LWPFQVINEG | GKPKVLVSYK | GENKAFYPEE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ISSMVLTKLK | ETAEAFLGHP | VTNAVITVPA | YFNDSQRQAT | KDAGVIAGLN | VLRIINEPTA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AAIAYGLDKG | GQGERHVLIF | DLGGGTFDVS | ILTIDDGIFE | VKATAGDTHL | GGEDFDNRLV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SHFVEEFKRK | HKKDISQNKR | AVRRLRTACE | RAKRTLSSST | QANLEIDSLY | EGIDFYTSIT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RARFEELCAD | LFRGTLEPVE | KALRDAKMDK | AKIHDIVLVG | GSTRIPKVQR | LLQDYFNGRD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LNKSINPDEA | VAYGAAVQAA | ILMGDKSEKV | QDLLLLDVAP | LSLGLETAGG | VMTALIKRNS |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TIPTKQTQIF | TTYSDNQPGV | LIQVYEGERA | MTKDNNLLGR | FDLTGIPPAP | RGVPQIEVTF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DIDANGILNV | TATDKSTGKV | NKITITNDKG | RLSKEEIERM | VLDAEKYKAE | DEVQREKIAA |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KNALESYAFN | MKSVVSDEGL | KGKISESDKN | KILDKCNELL | SWLEVNQLAE | KDEFDHKRKE |
| 610 | 620 | 630 | 640 | ||
| LEQMCNPIIT | KLYQGGCTGP | ACGTGYVPGR | PATGPTIEEV | D |