P34722
Gene name |
tpa-1 (B0545.1) |
Protein name |
Protein kinase C-like 1 |
Names |
PKC, Tetradecanoyl phorbol acetate-resistant protein 1 |
Species |
Caenorhabditis elegans |
KEGG Pathway |
|
EC number |
2.7.11.13: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
516-540 (Activation loop from InterPro)
Target domain |
375-634 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
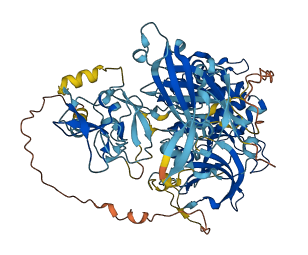
Autoinhibited structure

Activated structure

1 structures for P34722
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P34722-F1 | Predicted | AlphaFoldDB |
No variants for P34722
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P34722 | |||||
No associated diseases with P34722
7 regional properties for P34722
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Krueppel-associated box | 211 - 283 | IPR001909 |
| domain | SCAN domain | 37 - 149 | IPR003309 |
| domain | Zinc finger C2H2-type | 408 - 435 | IPR013087-1 |
| domain | Zinc finger C2H2-type | 436 - 463 | IPR013087-2 |
| domain | Zinc finger C2H2-type | 464 - 491 | IPR013087-3 |
| domain | Zinc finger C2H2-type | 492 - 519 | IPR013087-4 |
| domain | Zinc finger C2H2-type | 520 - 547 | IPR013087-5 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.13 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein kinase C activity | Calcium-dependent catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. |
| metal ion binding | Binding to a metal ion. |
| mitogen-activated protein kinase kinase kinase binding | Binding to a mitogen-activated protein kinase kinase kinase, a protein that can phosphorylate a MAP kinase kinase. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response to fungus | Reactions triggered in response to the presence of a fungus that act to protect the cell or organism. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of axon regeneration | Any process that activates, maintains or increases the rate of axon regeneration. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| response to wounding | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism. |
31 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P24583 | PKC1 | Protein kinase C-like 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| A1A4I4 | PKN1 | Serine/threonine-protein kinase N1 | Bos taurus (Bovine) | SS |
| A1Z7T0 | Pkn | Serine/threonine-protein kinase N | Drosophila melanogaster (Fruit fly) | SS |
| P83099 | Pkcdelta | Putative protein kinase C delta type homolog | Drosophila melanogaster (Fruit fly) | PR |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q96LW2 | RSKR | Ribosomal protein S6 kinase-related protein | Homo sapiens (Human) | PR |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P70268 | Pkn1 | Serine/threonine-protein kinase N1 | Mus musculus (Mouse) | SS |
| P23298 | Prkch | Protein kinase C eta type | Mus musculus (Mouse) | PR |
| Q8BWW9 | Pkn2 | Serine/threonine-protein kinase N2 | Mus musculus (Mouse) | SS |
| Q8K045 | Pkn3 | Serine/threonine-protein kinase N3 | Mus musculus (Mouse) | SS |
| P16054 | Prkce | Protein kinase C epsilon type | Mus musculus (Mouse) | PR |
| Q02111 | Prkcq | Protein kinase C theta type | Mus musculus (Mouse) | PR |
| P28867 | Prkcd | Protein kinase C delta type | Mus musculus (Mouse) | PR |
| Q63433 | Pkn1 | Serine/threonine-protein kinase N1 | Rattus norvegicus (Rat) | SS |
| Q64617 | Prkch | Protein kinase C eta type | Rattus norvegicus (Rat) | PR |
| P09216 | Prkce | Protein kinase C epsilon type | Rattus norvegicus (Rat) | PR |
| O08874 | Pkn2 | Serine/threonine-protein kinase N2 | Rattus norvegicus (Rat) | SS |
| P09215 | Prkcd | Protein kinase C delta type | Rattus norvegicus (Rat) | PR |
| Q9Y1J3 | pdk-1 | 3-phosphoinositide-dependent protein kinase 1 | Caenorhabditis elegans | SS |
| Q19266 | pkc-3 | Protein kinase C-like 3 | Caenorhabditis elegans | SS |
| P90980 | pkc-2 | Protein kinase C-like 2 | Caenorhabditis elegans | SS |
| Q9XTG7 | akt-2 | Serine/threonine-protein kinase akt-2 | Caenorhabditis elegans | SS |
| Q17941 | akt-1 | Serine/threonine-protein kinase akt-1 | Caenorhabditis elegans | PR |
| Q2L6W9 | sax-1 | Serine/threonine-protein kinase sax-1 | Caenorhabditis elegans | SS |
| A7MBL8 | pkn2 | Serine/threonine-protein kinase N2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAQAENACRL | KLLRADVPVD | LLPAGCSATD | LQPAVNVKEK | IEVNGESRLV | QKKKTLYPEW |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EKCWDTAVAE | GRILQIVLMF | NQTPVVEATM | RLEDIISKCK | SDAITHIWIN | TKPNGRILAQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TRHLKNAPDD | DHPVEDIMTS | RSNSGPGIQR | RRGAIKHARV | HEIRGHQFVA | TFFRQPHFCS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LCSDFMWGLN | KQGYQCQLCS | AAVHKKCHEK | VIMQCPGSAK | NTKETMALKE | RFKVDIPHRF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KTYNFKSPTF | CDHCGSMLYG | LFKQGLRCEV | CNVACHHKCE | RLMSNLCGVN | QKQLSEMYHE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IKRGTHATAS | CPPNIANLHL | NGETSKNNGS | LPNKLKNLFK | SHQYSVEEQK | ETDEYMDNIW |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GGGDGPVKKF | ALPHFNLLKV | LGKGSFGKVM | LVELKGKNEF | YAMKCLKKDV | ILEDDDTECT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YIERRVLILA | SQCPFLCQLF | CSFQTNEYLF | FVMEYLNGGD | LMHHIQQIKK | FDEARTRFYA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| CEIVVALQFL | HTNNIIYRDL | KLDNVLLDCD | GHIKLADFGM | AKTEMNRENG | MASTFCGTPD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| YISPEIIKGQ | LYNEAVDFWS | FGVLMYEMLV | GQSPFHGEGE | DELFDSILNE | RPYFPKTISK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EAAKCLSALF | DRNPNTRLGM | PECPDGPIRQ | HCFFRGVDWK | RFENRQVPPP | FKPNIKSNSD |
| 670 | 680 | 690 | 700 | ||
| ASNFDDDFTN | EKAALTPVHD | KNLLASIDPE | AFLNFSYTNP | HFSK |