P32485
Gene name |
HOG1 (SSK3, YLR113W, L2931, L9354.2) |
Protein name |
Mitogen-activated protein kinase HOG1 |
Names |
MAP kinase HOG1, High osmolarity glycerol response protein 1 |
Species |
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) |
KEGG Pathway |
sce:YLR113W |
EC number |
2.7.11.24: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
161-181 (Activation loop from InterPro)
Target domain |
23-302 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
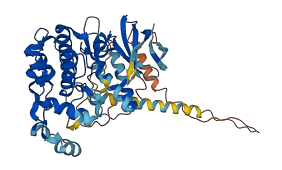
Autoinhibited structure

Activated structure

1 structures for P32485
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P32485-F1 | Predicted | AlphaFoldDB |
2 variants for P32485
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| s12-372525 | 302 | S>L | No | SGRP | |
| s12-372840 | 407 | N>S | No | SGRP |
No associated diseases with P32485
4 regional properties for P32485
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 23 - 302 | IPR000719 |
| conserved_site | Mitogen-activated protein (MAP) kinase, conserved site | 58 - 156 | IPR003527 |
| active_site | Serine/threonine-protein kinase, active site | 140 - 152 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 29 - 53 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.24 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| MAP kinase activity | Catalysis of the reaction: protein + ATP = protein phosphate + ADP. This reaction is the phosphorylation of proteins. Mitogen-activated protein kinase; a family of protein kinases that perform a crucial step in relaying signals from the plasma membrane to the nucleus. They are activated by a wide range of proliferation- or differentiation-inducing signals; activation is strong with agonists such as polypeptide growth factors and tumor-promoting phorbol esters, but weak (in most cell backgrounds) by stress stimuli. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| hyperosmotic response | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of detection of, or exposure to, a hyperosmotic environment, i.e. an environment with a higher concentration of solutes than the organism or cell. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of exit from mitosis | Any process involved in the inhibition of progression from anaphase/telophase (high mitotic CDK activity) to G1 (low mitotic CDK activity). |
| osmosensory signaling pathway | The series of molecular signals initiated in response to osmotic change. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of macroautophagy | Any process that modulates the frequency, rate or extent of macroautophagy. |
| regulation of nuclear cell cycle DNA replication | Any process that modulates the frequency, rate or extent of The DNA-dependent DNA replication that occurs in the nucleus of eukaryotic organisms as part of the cell cycle. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| stress-activated MAPK cascade | The series of molecular signals in which a stress-activated MAP kinase cascade relays a signal; MAP kinase cascades involve at least three protein kinase activities and culminate in the phosphorylation and activation of a MAP kinase. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P14681 | KSS1 | Mitogen-activated protein kinase KSS1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| P16892 | FUS3 | Mitogen-activated protein kinase FUS3 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| P41808 | SMK1 | Sporulation-specific mitogen-activated protein kinase SMK1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q00772 | SLT2 | Mitogen-activated protein kinase SLT2/MPK1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q3T0N5 | MAPK13 | Mitogen-activated protein kinase 13 | Bos taurus (Bovine) | SS |
| P79996 | MAPK9 | Mitogen-activated protein kinase 9 | Gallus gallus (Chicken) | PR |
| Q9N272 | MAPK13 | Mitogen-activated protein kinase 13 | Pan troglodytes (Chimpanzee) | SS |
| Q95NE7 | MAPK14 | Mitogen-activated protein kinase 14 | Pan troglodytes (Chimpanzee) | SS |
| P83100 | p38c | Putative mitogen-activated protein kinase 14C | Drosophila melanogaster (Fruit fly) | SS |
| O62618 | p38a | Mitogen-activated protein kinase p38a | Drosophila melanogaster (Fruit fly) | SS |
| O61443 | p38b | Mitogen-activated protein kinase p38b | Drosophila melanogaster (Fruit fly) | SS |
| P45983 | MAPK8 | Mitogen-activated protein kinase 8 | Homo sapiens (Human) | EV |
| P45984 | MAPK9 | Mitogen-activated protein kinase 9 | Homo sapiens (Human) | SS |
| P53779 | MAPK10 | Mitogen-activated protein kinase 10 | Homo sapiens (Human) | EV |
| P53778 | MAPK12 | Mitogen-activated protein kinase 12 | Homo sapiens (Human) | SS |
| O15264 | MAPK13 | Mitogen-activated protein kinase 13 | Homo sapiens (Human) | SS |
| Q15759 | MAPK11 | Mitogen-activated protein kinase 11 | Homo sapiens (Human) | SS |
| Q16539 | MAPK14 | Mitogen-activated protein kinase 14 | Homo sapiens (Human) | SS |
| Q91Y86 | Mapk8 | Mitogen-activated protein kinase 8 | Mus musculus (Mouse) | PR |
| Q9WTU6 | Mapk9 | Mitogen-activated protein kinase 9 | Mus musculus (Mouse) | PR |
| O08911 | Mapk12 | Mitogen-activated protein kinase 12 | Mus musculus (Mouse) | SS |
| Q9Z1B7 | Mapk13 | Mitogen-activated protein kinase 13 | Mus musculus (Mouse) | SS |
| Q9WUI1 | Mapk11 | Mitogen-activated protein kinase 11 | Mus musculus (Mouse) | SS |
| P47811 | Mapk14 | Mitogen-activated protein kinase 14 | Mus musculus (Mouse) | SS |
| P49186 | Mapk9 | Mitogen-activated protein kinase 9 | Rattus norvegicus (Rat) | PR |
| Q63538 | Mapk12 | Mitogen-activated protein kinase 12 | Rattus norvegicus (Rat) | SS |
| Q9WTY9 | Mapk13 | Mitogen-activated protein kinase 13 | Rattus norvegicus (Rat) | SS |
| P70618 | Mapk14 | Mitogen-activated protein kinase 14 | Rattus norvegicus (Rat) | SS |
| Q336X9 | MPK6 | Mitogen-activated protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q5J4W4 | MPK2 | Mitogen-activated protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84UI5 | MPK1 | Mitogen-activated protein kinase 1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10N20 | MPK5 | Mitogen-activated protein kinase 5 | Oryza sativa subsp. japonica (Rice) | SS |
| O44408 | kgb-1 | GLH-binding kinase 1 | Caenorhabditis elegans | PR |
| Q8MXI4 | pmk-2 | Mitogen-activated protein kinase pmk-2 | Caenorhabditis elegans | SS |
| Q17446 | pmk-1 | Mitogen-activated protein kinase pmk-1 | Caenorhabditis elegans | SS |
| Q39023 | MPK3 | Mitogen-activated protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39026 | MPK6 | Mitogen-activated protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39025 | MPK5 | Mitogen-activated protein kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GYQ5 | MPK12 | Mitogen-activated protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LMM5 | MPK11 | Mitogen-activated protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39024 | MPK4 | Mitogen-activated protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQQ9 | MPK13 | Mitogen-activated protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M1Z5 | MPK10 | Mitogen-activated protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O42376 | mapk12 | Mitogen-activated protein kinase 12 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9DGE2 | mapk14a | Mitogen-activated protein kinase 14A | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9DGE1 | mapk14b | Mitogen-activated protein kinase 14B | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTTNEEFIRT | QIFGTVFEIT | NRYNDLNPVG | MGAFGLVCSA | TDTLTSQPVA | IKKIMKPFST |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AVLAKRTYRE | LKLLKHLRHE | NLICLQDIFL | SPLEDIYFVT | ELQGTDLHRL | LQTRPLEKQF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VQYFLYQILR | GLKYVHSAGV | IHRDLKPSNI | LINENCDLKI | CDFGLARIQD | PQMTGYVSTR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YYRAPEIMLT | WQKYDVEVDI | WSAGCIFAEM | IEGKPLFPGK | DHVHQFSIIT | DLLGSPPKDV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| INTICSENTL | KFVTSLPHRD | PIPFSERFKT | VEPDAVDLLE | KMLVFDPKKR | ITAADALAHP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YSAPYHDPTD | EPVADAKFDW | HFNDADLPVD | TWRVMMYSEI | LDFHKIGGSD | GQIDISATFD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DQVAAATAAA | AQAQAQAQAQ | VQLNMAAHSH | NGAGTTGNDH | SDIAGGNKVS | DHVAANDTIT |
| 430 | |||||
| DYGNQAIQYA | NEFQQ |