P31976
Gene name |
EZR (VIL2) |
Protein name |
Ezrin |
Names |
Cytovillin, Villin-2, p81 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:281574 |
EC number |
|
Protein Class |
|
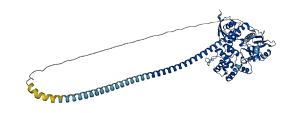
Descriptions
(Annotation based on sequence homology with P26038)
MSN is a moesin protein and belongs to the ezrin-radixin-moesin (ERM) protein family, directly involved in the cytoskeleton-membrane crosslinking. Functional N-terminal FERM domain of ERM attaches to the membrane by binding specific membrane proteins while the last 34 residues of the C-terminal tail domain bind actin filaments. The autoinhibitory domain is positions at the C-terminal tail domain of ERM, where FERM domain of ERM is bound tightly via phosphotyrosine binding (PTB), pleckstrin homology (PH), and Enabled/VASP Homology 1 (EVH1) domains, thus masking the binding sites for other molecules. ERM is activated through phosphorylation at Thr558 weakening the FERM/tail binding and, unmasks the binding sites of membrane protein and actin filaments in the presence of phospholipids.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P31976
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P31976-F1 | Predicted | AlphaFoldDB |
202 variants for P31976
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs454889138 | 3 | K>E | No | EVA | |
| rs442968750 | 4 | P>R | No | EVA | |
| rs526880533 | 6 | N>H | No | EVA | |
| rs459308463 | 7 | V>A | No | EVA | |
| rs477779712 | 7 | V>L | No | EVA | |
| rs473858781 | 9 | V>G | No | EVA | |
| rs461968275 | 10 | T>P | No | EVA | |
| rs443388565 | 11 | T>P | No | EVA | |
| rs476361909 | 12 | M>L | No | EVA | |
| rs451475350 | 12 | M>R | No | EVA | |
| rs472424564 | 13 | D>E | No | EVA | |
| rs433182388 | 13 | D>G | No | EVA | |
| rs453996301 | 14 | A>G | No | EVA | |
| rs435694123 | 15 | E>G | No | EVA | |
| rs468637647 | 16 | L>R | No | EVA | |
| rs450308350 | 17 | E>G | No | EVA | |
| rs438183456 | 21 | Q>H | No | EVA | |
| rs456687559 | 65 | V>G | No | EVA | |
| rs438356735 | 66 | S>A | No | EVA | |
| rs444855692 | 69 | E>A | No | EVA | |
| rs477843459 | 70 | V>D | No | EVA | |
| rs477843459 | 70 | V>G | No | EVA | |
| rs465743799 | 76 | L>P | No | EVA | |
| rs447316525 | 78 | F>L | No | EVA | |
| rs447316525 | 78 | F>V | No | EVA | |
| rs136241981 | 121 | T>P | No | EVA | |
| rs443376857 | 122 | A>G | No | EVA | |
| rs461789939 | 122 | A>T | No | EVA | |
| rs439449531 | 123 | V>A | No | EVA | |
| rs457947604 | 123 | V>L | No | EVA | |
| rs472599074 | 125 | L>M | No | EVA | |
| rs454051221 | 126 | G>R | No | EVA | |
| rs442100784 | 127 | S>Y | No | EVA | |
| rs475091351 | 128 | Y>D | No | EVA | |
| rs456773635 | 134 | F>V | No | EVA | |
| rs438185292 | 137 | Y>* | No | EVA | |
| rs471328799 | 142 | H>P | No | EVA | |
| rs452741268 | 143 | K>N | No | EVA | |
| rs434246467 | 146 | Y>S | No | EVA | |
| rs447197996 | 150 | E>K | No | EVA | |
| rs435299639 | 154 | P>L | No | EVA | |
| rs468105357 | 155 | Q>R | No | EVA | |
| rs433644116 | 180 | R>S | No | EVA | |
| rs436318335 | 181 | G>A | No | EVA | |
| rs454684279 | 181 | G>W | No | EVA | |
| rs524682677 | 229 | E>G | No | EVA | |
| rs438659284 | 235 | T>P | No | EVA | |
| rs434879013 | 254 | K>N | No | EVA | |
| rs432873579 | 266 | D>V | No | EVA | |
| rs451182660 | 266 | D>Y | No | EVA | |
| rs876277991 | 268 | V>G | No | EVA | |
| rs465695505 | 270 | Y>D | No | EVA | |
| rs453818524 | 270 | Y>S | No | EVA | |
| rs435382226 | 271 | A>P | No | EVA | |
| rs468295294 | 272 | P>S | No | EVA | |
| rs449987011 | 277 | N>I | No | EVA | |
| rs449987011 | 277 | N>T | No | EVA | |
| rs464590178 | 280 | I>S | No | EVA | |
| rs464590178 | 280 | I>T | No | EVA | |
| rs446060451 | 281 | L>R | No | EVA | |
| rs876110423 | 282 | Q>H | No | EVA | |
| rs876217302 | 282 | Q>R | No | EVA | |
| rs876033788 | 283 | L>H | No | EVA | |
| rs876033788 | 283 | L>R | No | EVA | |
| rs876485854 | 283 | L>V | No | EVA | |
| rs479148440 | 286 | G>V | No | EVA | |
| rs460647560 | 287 | N>Y | No | EVA | |
| rs448439222 | 291 | Y>F | No | EVA | |
| rs448439222 | 291 | Y>S | No | EVA | |
| rs481689890 | 292 | M>L | No | EVA | |
| rs463078906 | 295 | R>Q | No | EVA | |
| rs444762204 | 300 | I>M | No | EVA | |
| rs471086532 | 302 | V>G | No | EVA | |
| rs457623409 | 307 | A>S | No | EVA | |
| rs435247786 | 318 | L>R | No | EVA | |
| rs456248810 | 319 | E>Q | No | EVA | |
| rs479900921 | 320 | R>S | No | EVA | |
| rs460202168 | 332 | T>N | No | EVA | |
| rs481013918 | 349 | L>F | No | EVA | |
| rs462629996 | 351 | L>I | No | EVA | |
| rs470798220 | 355 | E>G | No | EVA | |
| rs452310860 | 358 | T>P | No | EVA | |
| rs440361620 | 362 | E>G | No | EVA | |
| rs437477209 | 412 | K>Q | No | EVA | |
| rs462085649 | 418 | A>P | No | EVA | |
| rs443774517 | 421 | L>P | No | EVA | |
| rs483093413 | 423 | E>D | No | EVA | |
| rs439821217 | 430 | L>R | No | EVA | |
| rs472741763 | 447 | L>V | No | EVA | |
| rs442260073 | 448 | R>P | No | EVA | |
| rs456310655 | 451 | E>Q | No | EVA | |
| rs470875884 | 454 | D>E | No | EVA | |
| rs444396455 | 454 | D>Y | No | EVA | |
| rs452342130 | 455 | D>A | No | EVA | |
| rs433979339 | 456 | L>R | No | EVA | |
| rs466963524 | 458 | K>Q | No | EVA | |
| rs454959317 | 459 | T>P | No | EVA | |
| rs469433816 | 460 | R>G | No | EVA | |
| rs465507483 | 461 | E>G | No | EVA | |
| rs477546295 | 461 | E>K | No | EVA | |
| rs447091461 | 462 | E>* | No | EVA | |
| rs460138560 | 462 | E>D | No | EVA | |
| rs478668695 | 462 | E>G | No | EVA | |
| rs447091461 | 462 | E>Q | No | EVA | |
| rs481110413 | 463 | L>R | No | EVA | |
| rs441722394 | 463 | L>V | No | EVA | |
| rs444257738 | 464 | H>D | No | EVA | |
| rs470738589 | 464 | H>L | No | EVA | |
| rs470738589 | 464 | H>P | No | EVA | |
| rs473421498 | 465 | L>R | No | EVA | |
| rs440256956 | 465 | L>V | No | EVA | |
| rs454894634 | 466 | V>A | No | EVA | |
| rs454894634 | 466 | V>G | No | EVA | |
| rs457232779 | 467 | M>I | No | EVA | |
| rs475697246 | 467 | M>L | No | EVA | |
| rs475697246 | 467 | M>V | No | EVA | |
| rs432553566 | 468 | T>A | No | EVA | |
| rs432553566 | 468 | T>P | No | EVA | |
| rs465545411 | 469 | A>P | No | EVA | |
| rs435054898 | 470 | P>A | No | EVA | |
| rs480977277 | 472 | P>T | No | EVA | |
| rs450592190 | 473 | P>Q | No | EVA | |
| rs450592190 | 473 | P>R | No | EVA | |
| rs458731887 | 474 | P>R | No | EVA | |
| rs473324468 | 475 | V>G | No | EVA | |
| rs440321324 | 475 | V>L | No | EVA | |
| rs457294276 | 476 | Y>* | No | EVA | |
| rs475560158 | 476 | Y>C | No | EVA | |
| rs442788389 | 476 | Y>D | No | EVA | |
| rs475560158 | 476 | Y>F | No | EVA | |
| rs442788389 | 476 | Y>H | No | EVA | |
| rs475560158 | 476 | Y>S | No | EVA | |
| rs432414925 | 477 | E>Q | No | EVA | |
| rs472004429 | 478 | P>A | No | EVA | |
| rs453477338 | 479 | V>G | No | EVA | |
| rs468102614 | 481 | Y>F | No | EVA | |
| rs449560656 | 482 | H>P | No | EVA | |
| rs435999604 | 487 | P>Q | No | EVA | |
| rs450655236 | 491 | G>D | No | EVA | |
| rs468938324 | 491 | G>R | No | EVA | |
| rs477075599 | 495 | S>G | No | EVA | |
| rs458725918 | 495 | S>N | No | EVA | |
| rs458725918 | 495 | S>T | No | EVA | |
| rs446624384 | 497 | E>G | No | EVA | |
| rs461253656 | 498 | L>P | No | EVA | |
| rs479613704 | 498 | L>V | No | EVA | |
| rs482038976 | 499 | S>A | No | EVA | |
| rs463564786 | 500 | S>R | No | EVA | |
| rs471905285 | 501 | E>G | No | EVA | |
| rs438909078 | 501 | E>Q | No | EVA | |
| rs453296760 | 502 | G>D | No | EVA | |
| rs453296760 | 502 | G>V | No | EVA | |
| rs474383051 | 503 | I>S | No | EVA | |
| rs456046993 | 508 | N>T | No | EVA | |
| rs468997577 | 513 | I>M | No | EVA | |
| rs437478177 | 513 | I>S | No | EVA | |
| rs456922746 | 514 | T>P | No | EVA | |
| rs456922746 | 514 | T>S | No | EVA | |
| rs432102381 | 515 | E>D | No | EVA | |
| rs465073485 | 516 | A>P | No | EVA | |
| rs446458256 | 519 | N>T | No | EVA | |
| rs474458830 | 528 | T>N | No | EVA | |
| rs443989821 | 533 | L>M | No | EVA | |
| rs476887626 | 533 | L>Q | No | EVA | |
| rs451980401 | 536 | A>D | No | EVA | |
| rs451980401 | 536 | A>G | No | EVA | |
| rs431959078 | 538 | D>H | No | EVA | |
| rs452999394 | 539 | E>A | No | EVA | |
| rs434463641 | 539 | E>D | No | EVA | |
| rs464896167 | 539 | E>K | No | EVA | |
| rs467450118 | 541 | K>R | No | EVA | |
| rs448987599 | 542 | R>P | No | EVA | |
| rs436919977 | 543 | T>A | No | EVA | |
| rs469913493 | 545 | N>K | No | EVA | |
| rs445065265 | 547 | I>T | No | EVA | |
| rs459674806 | 548 | I>M | No | EVA | |
| rs478268602 | 548 | I>N | No | EVA | |
| rs447786895 | 554 | R>S | No | EVA | |
| rs480770930 | 555 | Q>K | No | EVA | |
| rs462382529 | 556 | G>V | No | EVA | |
| rs443849064 | 558 | D>A | No | EVA | |
| rs443849064 | 558 | D>V | No | EVA | |
| rs458450683 | 560 | Y>* | No | EVA | |
| rs476750736 | 560 | Y>D | No | EVA | |
| rs439856609 | 561 | K>E | No | EVA | |
| rs452807632 | 562 | T>P | No | EVA | |
| rs455226591 | 565 | Q>K | No | EVA | |
| rs436981683 | 565 | Q>P | No | EVA | |
| rs469776461 | 566 | I>S | No | EVA | |
| rs480571436 | 568 | Q>H | No | EVA | |
| rs447577339 | 568 | Q>K | No | EVA | |
| rs462243409 | 570 | N>I | No | EVA | |
| rs450181273 | 573 | Q>R | No | EVA | |
| rs458258041 | 574 | R>P | No | EVA | |
| rs483221173 | 574 | R>S | No | EVA | |
| rs439910139 | 576 | D>H | No | EVA | |
| rs460814389 | 578 | F>V | No | EVA | |
| rs440797519 | 579 | E>K | No | EVA | |
| rs440797519 | 579 | E>Q | No | EVA | |
| rs473574399 | 581 | M>R | No | EVA | |
| rs455291996 | 582 | M>R | No | EVA | |
| rs436846576 | 582 | M>S | No | EVA |
No associated diseases with P31976
10 regional properties for P31976
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 5 - 295 | IPR000299 |
| domain | Ezrin/radixin/moesin, C-terminal | 506 - 581 | IPR011259 |
| domain | FERM, N-terminal | 9 - 68 | IPR018979 |
| domain | FERM, C-terminal PH-like domain | 210 - 299 | IPR018980 |
| conserved_site | FERM conserved site | 58 - 88 | IPR019747-1 |
| conserved_site | FERM conserved site | 176 - 205 | IPR019747-2 |
| domain | FERM central domain | 92 - 206 | IPR019748 |
| domain | Band 4.1 domain | 1 - 206 | IPR019749 |
| domain | ERM family, FERM domain C-lobe | 200 - 296 | IPR041789 |
| domain | Ezrin/radixin/moesin, alpha-helical domain | 337 - 456 | IPR046810 |
Functions
15 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| actin filament | A filamentous structure formed of a two-stranded helical polymer of the protein actin and associated proteins. Actin filaments are a major component of the contractile apparatus of skeletal muscle and the microfilaments of the cytoskeleton of eukaryotic cells. The filaments, comprising polymerized globular actin molecules, appear as flexible structures with a diameter of 5-9 nm. They are organized into a variety of linear bundles, two-dimensional networks, and three dimensional gels. In the cytoskeleton they are most highly concentrated in the cortex of the cell just beneath the plasma membrane. |
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| apical part of cell | The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue. |
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| basolateral plasma membrane | The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| ciliary basal body | A membrane-tethered, short cylindrical array of microtubules and associated proteins found at the base of a eukaryotic cilium (also called flagellum) that is similar in structure to a centriole and derives from it. The cilium basal body is the site of assembly and remodelling of the cilium and serves as a nucleation site for axoneme growth. As well as anchoring the cilium, it is thought to provide a selective gateway regulating the entry of ciliary proteins and vesicles by intraflagellar transport. |
| extrinsic component of membrane | The component of a membrane consisting of gene products and protein complexes that are loosely bound to one of its surfaces, but not integrated into the hydrophobic region. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| microvillus | Thin cylindrical membrane-covered projections on the surface of an animal cell containing a core bundle of actin filaments. Present in especially large numbers on the absorptive surface of intestinal cells. |
| microvillus membrane | The portion of the plasma membrane surrounding a microvillus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
| uropod | A membrane projection with related cytoskeletal components at the trailing edge of a cell in the process of migrating or being activated, found on the opposite side of the cell from the leading edge or immunological synapse, respectively. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| cell adhesion molecule binding | Binding to a cell adhesion molecule. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament bundle assembly | The assembly of actin filament bundles; actin filaments are on the same axis but may be oriented with the same or opposite polarities and may be packed with different levels of tightness. |
| establishment or maintenance of apical/basal cell polarity | Any cellular process that results in the specification, formation or maintenance polarization of a cell's architecture along its apical/basal axis so that the apical and basal regions of the cell have different membrane, extracellular matrix and sub-membrane cellular components. |
| positive regulation of early endosome to late endosome transport | Any process that activates or increases the frequency, rate or extent of early endosome to late endosome transport. |
| positive regulation of protein localization to early endosome | Any process that activates or increases the frequency, rate or extent of protein localization to early endosome. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of organelle assembly | Any process that modulates the frequency, rate or extent of organelle assembly. |
21 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2HJ49 | MSN | Moesin | Bos taurus (Bovine) | PR |
| Q32LP2 | RDX | Radixin | Bos taurus (Bovine) | SS |
| Q9PU45 | RDX | Radixin | Gallus gallus (Chicken) | PR |
| Q24564 | Mer | Moesin/ezrin/radixin homolog 2 | Drosophila melanogaster (Fruit fly) | SS |
| P46150 | Moe | Moesin/ezrin/radixin homolog 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q3KP66 | INAVA | Innate immunity activator protein | Homo sapiens (Human) | PR |
| P35240 | NF2 | Merlin | Homo sapiens (Human) | EV |
| P35241 | RDX | Radixin | Homo sapiens (Human) | SS |
| P15311 | EZR | Ezrin | Homo sapiens (Human) | EV |
| P26038 | MSN | Moesin | Homo sapiens (Human) | EV |
| A2AD83 | Frmd7 | FERM domain-containing protein 7 | Mus musculus (Mouse) | PR |
| P26043 | Rdx | Radixin | Mus musculus (Mouse) | SS |
| P46662 | Nf2 | Merlin | Mus musculus (Mouse) | SS |
| P26040 | Ezr | Ezrin | Mus musculus (Mouse) | SS |
| P26041 | Msn | Moesin | Mus musculus (Mouse) | SS |
| P26042 | MSN | Moesin | Sus scrofa (Pig) | PR |
| P26044 | RDX | Radixin | Sus scrofa (Pig) | SS |
| Q63648 | Nf2 | Merlin | Rattus norvegicus (Rat) | SS |
| O35763 | Msn | Moesin | Rattus norvegicus (Rat) | SS |
| P31977 | Ezr | Ezrin | Rattus norvegicus (Rat) | SS |
| Q6Q413 | nf2b | NF2, moesin-ezrin-radixin-like | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPKPINVRVT | TMDAELEFAI | QPNTTGKQLF | DQVVKTIGLR | EVWYFGLQYV | DNKGFPTWLK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LDKKVSAQEV | RKESPLQFKF | RAKFYPEDVA | EELIQDITQK | LFFLQVKEGI | LSDEIYCPPE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TAVLLGSYAV | QAKFGDYNKE | LHKAGYLGSE | RLIPQRVMDQ | HKLTRDQWED | RIQVWHAEHR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GMLKDSAMLE | YLKIAQDLEM | YGINYFEIKN | KKGTDLWLGV | DALGLNIYEK | DDKLTPKIGF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PWSEIRNISF | NDKKFVIKPI | DKKAPDFVFY | APRLRINKRI | LQLCMGNHEL | YMRRRKPDTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EVQQMKAQAR | EEKHQKQLER | QQLETEKKRR | ETVEREKEQM | MREKEELMLR | LQDYEEKTRK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AEKELSDQIQ | RALKLEEERK | RAQEEAGRLE | ADRLAALRAK | EELERQAADQ | IKSQEQLATE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LAEYTAKIAL | LEEARRRKEN | EVEEWQLRAK | EAQDDLVKTR | EELHLVMTAP | PPPPVYEPVN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YHVHEGPQEE | GTELSAELSS | EGILDDRNEE | KRITEAEKNE | RVQRQLMTLT | SELSQARDEN |
| 550 | 560 | 570 | 580 | ||
| KRTHNDIIHN | ENMRQGRDKY | KTLRQIRQGN | TKQRIDEFEA | M |