P31946
Gene name |
YWHAB |
Protein name |
14-3-3 protein beta/alpha |
Names |
Protein 1054, Protein kinase C inhibitor protein 1, KCIP-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:7529 |
EC number |
|
Protein Class |
|
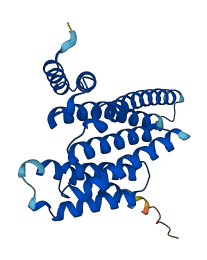
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

16 structures for P31946
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2BQ0 | X-ray | 250 A | A/B | 2-239 | PDB |
| 2C23 | X-ray | 265 A | A | 2-239 | PDB |
| 4DNK | X-ray | 220 A | A/B | 1-246 | PDB |
| 5N10 | X-ray | 160 A | A/B | 1-246 | PDB |
| 6A5Q | X-ray | 200 A | A/B/C | 1-246 | PDB |
| 6BYK | X-ray | 300 A | A/B/C/D | 3-232 | PDB |
| 6GN0 | X-ray | 324 A | A/B/C/D | 1-239 | PDB |
| 6GN8 | X-ray | 234 A | A/B | 1-234 | PDB |
| 6GNJ | X-ray | 324 A | A/B | 1-234 | PDB |
| 6GNK | X-ray | 255 A | A/B | 1-234 | PDB |
| 6GNN | X-ray | 379 A | A | 1-239 | PDB |
| 6HEP | X-ray | 186 A | A/B/C/D | 1-232 | PDB |
| 8DP5 | EM | 310 A | C | 1-246 | PDB |
| 8EQ8 | X-ray | 150 A | A/B | 1-239 | PDB |
| 8EQH | X-ray | 190 A | A/B | 1-239 | PDB |
| AF-P31946-F1 | Predicted | AlphaFoldDB |
120 variants for P31946
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs748780359 CA9872543 |
3 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA409130411 rs1372131336 |
4 | D>G | No |
ClinGen TOPMed |
|
|
rs1298127050 CA409130418 |
5 | K>E | No |
ClinGen TOPMed |
|
| TCGA novel | 6 | S>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA409130472 rs1394516879 |
10 | Q>R | No |
ClinGen gnomAD |
|
|
rs1411889702 CA409130484 |
11 | K>R | No |
ClinGen gnomAD |
|
|
CA315483813 rs745844801 |
15 | A>S | No |
ClinGen gnomAD |
|
|
CA409130530 rs745844801 |
15 | A>T | No |
ClinGen gnomAD |
|
|
CA409130570 rs1232040212 |
18 | A>T | No |
ClinGen gnomAD |
|
|
CA409130596 rs1170519701 |
20 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1227070320 CA409130605 |
21 | Y>F | No |
ClinGen gnomAD |
|
|
rs775702775 CA9872548 |
22 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs778607409 CA315483825 |
25 | A>V | No |
ClinGen Ensembl |
|
|
CA9872549 rs760945751 |
26 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1205402379 CA409130654 |
26 | A>P | No |
ClinGen gnomAD |
|
|
CA9872550 rs768860792 |
28 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs923525230 CA315483833 |
29 | K>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA409130700 rs1427708340 |
31 | V>F | No |
ClinGen gnomAD |
|
|
rs1168134597 CA409130707 |
32 | T>S | No |
ClinGen gnomAD |
|
| TCGA novel | 33 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs761774775 CA9872552 |
33 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs765793345 CA9872553 |
35 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA9872555 rs763376032 |
36 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs1412953854 CA409130755 |
36 | H>Y | No |
ClinGen gnomAD |
|
|
rs933285246 CA315483844 |
39 | S>Y | No |
ClinGen TOPMed |
|
|
rs1380617936 CA409130802 |
40 | N>D | No |
ClinGen gnomAD |
|
|
rs766591292 CA409130808 |
40 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755875208 CA9872558 |
46 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs753424390 CA9872560 |
47 | S>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 51 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA409130997 rs1237546287 |
54 | V>I | No |
ClinGen gnomAD |
|
|
CA409131039 rs779889895 |
56 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA9872565 rs779889895 |
56 | A>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 56 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA409131058 rs1249701757 |
57 | R>H | No |
ClinGen gnomAD |
|
|
rs11544163 CA315483864 |
57 | R>S | No |
ClinGen Ensembl |
|
|
rs1278496171 CA409131069 |
58 | R>C | No |
ClinGen TOPMed |
|
|
CA315483866 rs11556823 |
58 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs1601096343 CA409131080 |
59 | S>P | No |
ClinGen Ensembl |
|
|
CA409131140 COSM443878 rs1449802993 |
62 | R>C | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1236709695 CA409131144 |
62 | R>H | No |
ClinGen TOPMed |
|
|
CA9872567 rs768913881 |
64 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA9872566 rs746774284 |
64 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA409131235 rs1349100665 |
67 | I>T | No |
ClinGen TOPMed |
|
|
CA9872568 rs776783216 |
69 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA9872569 rs761980766 |
70 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1359792405 CA409131341 |
73 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA9872570 rs556782816 |
74 | N>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA9872573 rs766965462 |
76 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9872574 rs142757633 |
77 | K>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 79 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs201036178 CA315483886 |
79 | Q>R | No |
ClinGen 1000Genomes |
|
|
rs947477879 CA315483888 |
80 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1220580864 CA409131442 |
80 | M>L | No |
ClinGen gnomAD |
|
|
rs763872029 CA9872576 |
82 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1330178791 CA409131499 |
85 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1014116195 CA315483894 |
85 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA9872578 rs756805731 |
87 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA9872579 rs764712419 |
89 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1363076773 CA409131562 |
90 | A>V | No |
ClinGen TOPMed |
|
|
CA9872580 rs749944173 |
97 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409131694 rs758306820 |
98 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758306820 CA9872581 |
98 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| VAR_064762 | 99 | V>I | found in a renal cell carcinoma sample; somatic mutation [UniProt] | No | UniProt |
|
CA9872602 rs751428560 |
101 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409132192 rs1568636863 |
103 | L>S | No |
ClinGen Ensembl |
|
|
CA409132200 rs1407604864 |
104 | D>N | No |
ClinGen gnomAD |
|
|
CA9872606 rs535043251 |
109 | P>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA315484337 rs990395014 |
109 | P>T | No |
ClinGen Ensembl |
|
|
CA315484345 rs955232966 |
110 | N>D | No |
ClinGen TOPMed |
|
|
CA409132272 rs1358220876 |
110 | N>K | No |
ClinGen gnomAD |
|
|
rs151038182 CA9872607 |
110 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs749361701 CA409132279 |
111 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs749361701 CA9872608 |
111 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1437521308 CA409132284 |
112 | T>A | No |
ClinGen gnomAD |
|
|
rs1437521308 CA409132282 |
112 | T>P | No |
ClinGen gnomAD |
|
|
rs1170885547 CA409132322 |
115 | E>G | No |
ClinGen gnomAD |
|
|
CA409132342 rs1203257313 |
117 | K>E | No |
ClinGen gnomAD |
|
| TCGA novel | 117 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9872610 rs775014630 |
117 | K>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9872612 rs772702305 |
123 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA315484369 rs6031861 |
128 | F>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1226064691 CA409132557 |
131 | L>V | No |
ClinGen TOPMed |
|
|
CA9872614 rs761079143 |
135 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA9872615 rs764818610 |
136 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs143477601 CA9872617 |
139 | N>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs765814877 CA9872618 |
141 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs1366272647 CA409132893 |
142 | T>I | No |
ClinGen gnomAD |
|
|
CA409132898 rs767389512 |
143 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA9872638 rs767389512 |
143 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs2664553 CA315484690 |
144 | V>E | No |
ClinGen TOPMed |
|
|
CA9872639 rs752616742 |
145 | S>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 150 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 154 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 162 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1035934124 CA315484708 |
166 | H>Q | No |
ClinGen Ensembl |
|
|
rs779254957 CA9872644 |
168 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1218297734 CA409133348 COSM3841089 |
169 | R>C | Variant assessed as Somatic; impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1190977283 CA409133350 |
169 | R>H | No |
ClinGen gnomAD |
|
|
CA409133436 rs1249016960 |
173 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 180 | Y>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs758462959 CA9872646 |
181 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1157941195 CA409133831 |
186 | S>F | No |
ClinGen gnomAD |
|
|
rs1386279474 CA409133858 |
187 | P>S | No |
ClinGen gnomAD |
|
|
rs1386279474 CA409133841 |
187 | P>T | No |
ClinGen gnomAD |
|
|
rs747489177 CA9872648 |
188 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA409134061 rs1208129243 |
192 | S>I | No |
ClinGen gnomAD |
|
|
rs867085274 CA315484724 |
194 | A>T | No |
ClinGen Ensembl |
|
|
rs146766895 CA9872649 |
196 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs750604097 CA9872662 |
197 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1230801307 CA409134580 |
202 | I>V | No |
ClinGen gnomAD |
|
|
rs1321509405 CA409135055 |
203 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs374058745 CA9872664 |
207 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA9872667 rs781631165 |
217 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA409135675 rs1329415749 |
231 | T>S | No |
ClinGen TOPMed |
|
|
CA9872695 rs768214899 |
232 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs137964982 CA9872698 |
238 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 240 | D>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9872700 rs762630587 |
241 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA409135809 rs1464275498 |
241 | A>V | No |
ClinGen gnomAD |
No associated diseases with P31946
3 regional properties for P31946
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | 14-3-3 protein, conserved site | 43 - 53 | IPR023409-1 |
| conserved_site | 14-3-3 protein, conserved site | 213 - 232 | IPR023409-2 |
| domain | 14-3-3 domain | 5 - 244 | IPR023410 |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| melanosome | A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| vacuolar membrane | The lipid bilayer surrounding the vacuole and separating its contents from the cytoplasm of the cell. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| histone deacetylase binding | Binding to histone deacetylase. |
| identical protein binding | Binding to an identical protein or proteins. |
| phosphoprotein binding | Binding to a phosphorylated protein. |
| phosphoserine residue binding | Binding to a phosphorylated serine residue within a protein. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a protein kinase, an enzyme which phosphorylates a protein. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cytoplasmic sequestering of protein | The selective interaction of a protein with specific molecules in the cytoplasm, thereby inhibiting its transport into other areas of the cell. |
| negative regulation of G protein-coupled receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of G protein-coupled receptor signaling pathway. |
| negative regulation of protein dephosphorylation | Any process the stops, prevents, or reduces the frequency, rate or extent of removal of phosphate groups from a protein. |
| positive regulation of catalytic activity | Any process that activates or increases the activity of an enzyme. |
| protein localization | Any process in which a protein is transported to, or maintained in, a specific location. |
| protein targeting | The process of targeting specific proteins to particular regions of the cell, typically membrane-bounded subcellular organelles. Usually requires an organelle specific protein sequence motif. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
37 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | Homo sapiens (Human) | PR |
| P27348 | YWHAQ | 14-3-3 protein theta | Homo sapiens (Human) | PR |
| P61981 | YWHAG | 14-3-3 protein gamma | Homo sapiens (Human) | PR |
| Q04917 | YWHAH | 14-3-3 protein eta | Homo sapiens (Human) | PR |
| P49106 | GRF1 | 14-3-3-like protein GF14-6 | Zea mays (Maize) | PR |
| P68510 | Ywhah | 14-3-3 protein eta | Mus musculus (Mouse) | PR |
| P63101 | Ywhaz | 14-3-3 protein zeta/delta | Mus musculus (Mouse) | PR |
| P93784 | HOX | 14-3-3-like protein 16R | Solanum tuberosum (Potato) | PR |
| Q41418 | 14-3-3-like protein | Solanum tuberosum (Potato) | PR | |
| P63102 | Ywhaz | 14-3-3 protein zeta/delta | Rattus norvegicus (Rat) | PR |
| Q6EUP4 | GF14E | 14-3-3-like protein GF14-E | Oryza sativa subsp japonica (Rice) | PR |
| Q06967 | GF14F | 14-3-3-like protein GF14-F | Oryza sativa subsp japonica (Rice) | PR |
| Q7XTE8 | GF14B | 14-3-3-like protein GF14-B | Oryza sativa subsp japonica (Rice) | PR |
| Q6ZKC0 | GF14C | 14-3-3-like protein GF14-C | Oryza sativa subsp japonica (Rice) | PR |
| Q84J55 | GF14A | 14-3-3-like protein GF14-A | Oryza sativa subsp japonica (Rice) | PR |
| Q2R2W2 | GF14D | 14-3-3-like protein GF14-D | Oryza sativa subsp japonica (Rice) | PR |
| Q2R1D5 | GF14H | Putative 14-3-3-like protein GF14-H | Oryza sativa subsp japonica (Rice) | PR |
| Q96450 | GF14A | 14-3-3-like protein A | Glycine max (Soybean) (Glycine hispida) | PR |
| Q96453 | GF14D | 14-3-3-like protein D | Glycine max (Soybean) (Glycine hispida) | PR |
| Q96452 | GF14C | 14-3-3-like protein C | Glycine max (Soybean) (Glycine hispida) | PR |
| Q9C5W6 | GRF12 | 14-3-3-like protein GF14 iota | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9S9Z8 | GRF11 | 14-3-3-like protein GF14 omicron | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P46077 | GRF4 | 14-3-3-like protein GF14 phi | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q01525 | GRF2 | 14-3-3-like protein GF14 omega | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q96300 | GRF7 | 14-3-3-like protein GF14 nu | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42645 | GRF5 | 14-3-3-like protein GF14 upsilon | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42644 | GRF3 | 14-3-3-like protein GF14 psi | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q96299 | GRF9 | 14-3-3-like protein GF14 mu | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42652 | TFT4 | 14-3-3 protein 4 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93212 | TFT7 | 14-3-3 protein 7 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93209 | TFT3 | 14-3-3 protein 3 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93207 | TFT10 | 14-3-3 protein 10 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93211 | TFT6 | 14-3-3 protein 6 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93206 | TFT1 | 14-3-3 protein 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93208 | TFT2 | 14-3-3 protein 2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93213 | TFT8 | 14-3-3 protein 8 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93214 | TFT9 | 14-3-3 protein 9 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTMDKSELVQ | KAKLAEQAER | YDDMAAAMKA | VTEQGHELSN | EERNLLSVAY | KNVVGARRSS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| WRVISSIEQK | TERNEKKQQM | GKEYREKIEA | ELQDICNDVL | ELLDKYLIPN | ATQPESKVFY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LKMKGDYFRY | LSEVASGDNK | QTTVSNSQQA | YQEAFEISKK | EMQPTHPIRL | GLALNFSVFY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YEILNSPEKA | CSLAKTAFDE | AIAELDTLNE | ESYKDSTLIM | QLLRDNLTLW | TSENQGDEGD |
| AGEGEN |