P31751
Gene name |
AKT2 |
Protein name |
RAC-beta serine/threonine-protein kinase |
Names |
Protein kinase Akt-2, Protein kinase B beta, PKB beta, RAC-PK-beta |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:208 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
RIBOSOMAL PROTEIN S6 KINASE (PTHR24351) |
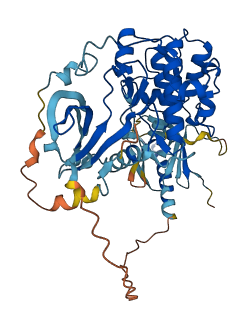
Descriptions
Akt is a serine/threonine kinase that belongs to a group of proteins called the AGC superfamily. It is involved in various biological responses through the phosphatidylinositol 3-kinase (PI3K) signal transduction pathway. Akt2, in particular, is regulated by different regions of the protein.
The N-terminal PH domain of Akt2 plays a regulatory role in its activation. Phosphorylation of Ser474 in the C-terminal regulatory domain is also necessary for full activation of Akt2. These modifications help activate the kinase and enable it to carry out its functions.
The linker region between the N-lobe and C-lobe of Akt2 occupies the ATP binding site. Additionally, the activation loop within Akt2 aids in autoinhibition by blocking the binding of peptide substrates, while still allowing access to the ATP binding site.
Autoinhibitory domains (AIDs)
Target domain |
143-481 (Kinase domain) |
Relief mechanism |
PTM, Ligand binding |
Assay |
|
Target domain |
143-481 (Kinase domain) |
Relief mechanism |
PTM |
Assay |
Mutagenesis experiment, Structural analysis |
Accessory elements
292-315 (Activation loop from InterPro)
Target domain |
156-478 (Catalytic domain of the Serine/Threonine Kinase, Protein Kinase B beta, also called Akt2) |
Relief mechanism |
|
Assay |
|
References
- Huang X et al. (2003) "Crystal structure of an inactive Akt2 kinase domain", Structure (London, England : 1993), 11, 21-30
- Truebestein L et al. (2021) "Structure of autoinhibited Akt1 reveals mechanism of PIP(3)-mediated activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Lučić I et al. (2018) "Conformational sampling of membranes by Akt controls its activation and inactivation", Proceedings of the National Academy of Sciences of the United States of America, 115, E3940-E3949
Autoinhibited structure
Activated structure

19 structures for P31751
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1GZK | X-ray | 230 A | A | 146-460 | PDB |
| 1GZN | X-ray | 250 A | A | 146-480 | PDB |
| 1GZO | X-ray | 275 A | A | 146-460 | PDB |
| 1MRV | X-ray | 280 A | A | 143-481 | PDB |
| 1MRY | X-ray | 280 A | A | 143-481 | PDB |
| 1O6K | X-ray | 170 A | A | 146-481 | PDB |
| 1O6L | X-ray | 160 A | A | 146-467 | PDB |
| 1P6S | NMR | - | A | 1-111 | PDB |
| 2JDO | X-ray | 180 A | A | 146-467 | PDB |
| 2JDR | X-ray | 230 A | A | 146-467 | PDB |
| 2UW9 | X-ray | 210 A | A | 146-467 | PDB |
| 2X39 | X-ray | 193 A | A | 146-467 | PDB |
| 2XH5 | X-ray | 272 A | A | 146-479 | PDB |
| 3D0E | X-ray | 200 A | A/B | 146-480 | PDB |
| 3E87 | X-ray | 230 A | A/B | 146-480 | PDB |
| 3E88 | X-ray | 250 A | A/B | 146-480 | PDB |
| 3E8D | X-ray | 270 A | A/B | 146-480 | PDB |
| 8Q61 | X-ray | 232 A | A | 1-481 | PDB |
| AF-P31751-F1 | Predicted | AlphaFoldDB |
282 variants for P31751
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1976263420 RCV001335851 |
15 | R>H | Type 2 diabetes mellitus [ClinVar] | Yes |
ClinVar dbSNP |
|
CA128662 VAR_067309 RCV000022676 rs387906659 COSM159009 |
17 | E>K | Hypoinsulinemic hypoglycemia and body hemihypertrophy breast HIHGHH; exhibits plasma membrane localization in serum-starved cells and produced inappropriate tonic nuclear exclusion of FOXO1 in preadipocytes [ClinVar, Cosmic, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl dbSNP |
|
VAR_067310 rs121434593 RCV000015016 CA123659 |
274 | R>H | NIDDM; associated with typical metabolic dyslipidemia with elevated fastin triglyceride, high VLDL triglyceride/cholesterol ratios, low HDL cholesterol levels and high small dense LDL levels; de novo lipogenesis and liver fat are also significantly elevated in this subject Type 2 diabetes mellitus [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
|
RCV000423006 rs1057519754 CA16602506 |
302 | S>G | Neoplasm of the large intestine [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
COSM3960070 rs778561687 RCV000441898 CA16602505 |
371 | R>H | lung Non-small cell lung carcinoma [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar ExAC dbSNP gnomAD |
|
CA9441513 rs771065673 RCV000624382 |
416 | D>N | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA405874606 rs1227207335 |
2 | N>S | No |
ClinGen gnomAD |
|
|
CA405874554 rs1386086141 |
6 | V>F | No |
ClinGen gnomAD |
|
|
rs199748431 CA405874542 |
7 | I>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs199748431 CA9442020 |
7 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1405458685 CA405874523 |
8 | K>R | No |
ClinGen gnomAD |
|
|
rs748173515 CA9442019 |
9 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1476241262 CA405874462 |
14 | K>T | No |
ClinGen gnomAD |
|
|
CA9442017 rs781407242 |
15 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs387906659 CA308392149 |
17 | E>* | No |
ClinGen Ensembl |
|
|
rs1438443076 CA405873281 |
19 | I>F | No |
ClinGen TOPMed |
|
|
CA405873280 rs1600058964 |
19 | I>T | No |
ClinGen Ensembl |
|
|
rs1272565080 CA405873213 |
24 | P>L | No |
ClinGen gnomAD |
|
|
rs1185991201 CA405873218 |
24 | P>S | No |
ClinGen TOPMed |
|
|
CA9441993 rs746989021 |
25 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1371021819 CA405873198 |
26 | Y>S | No |
ClinGen gnomAD |
|
|
CA9441992 rs779811637 |
31 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1360665207 CA405873136 |
32 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1197293098 CA405872995 |
41 | R>K | No |
ClinGen gnomAD |
|
|
CA9441986 rs763646270 |
45 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA9441985 rs201917021 |
49 | L>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9441984 rs201917021 |
49 | L>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs184042322 CA9441982 |
50 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs184042322 CA9441981 |
50 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA16040265 rs201145928 |
51 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs770250877 CA9441980 |
51 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770250877 CA405872815 |
51 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1407888862 CA405872708 |
57 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1380555769 CA405871665 |
61 | Q>R | No |
ClinGen gnomAD |
|
|
CA9441947 rs747803532 |
66 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1410236931 CA405871546 |
68 | P>L | No |
ClinGen gnomAD |
|
|
CA405871540 rs1287086134 |
69 | R>* | No |
ClinGen TOPMed |
|
|
COSM996631 CA405871534 rs1485745630 |
69 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA405871530 rs1170693725 |
70 | P>A | No |
ClinGen gnomAD |
|
|
rs200272953 CA9441945 |
71 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs200272953 CA405871507 |
71 | N>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs765768303 CA9441943 |
72 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA405871491 rs765768303 |
72 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA405871482 rs1190606056 |
73 | F>V | No |
ClinGen gnomAD |
|
|
CA9441941 rs754309980 |
74 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405871469 rs754309980 |
74 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405871326 rs1393896368 |
85 | E>K | No |
ClinGen TOPMed |
|
|
CA405871278 rs1489834693 |
88 | F>L | No |
ClinGen gnomAD |
|
|
CA405871240 rs1160079555 |
90 | V>M | No |
ClinGen TOPMed |
|
|
CA405871223 rs1205519703 |
91 | D>Y | No |
ClinGen gnomAD |
|
|
rs764512283 CA9441940 |
92 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA308390831 rs965617770 |
93 | P>L | No |
ClinGen Ensembl |
|
|
rs374073998 CA9441938 |
95 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA405869652 rs1434565896 |
97 | E>G | No |
ClinGen gnomAD |
|
|
CA9441916 rs369128708 |
100 | M>L | No |
ClinGen ESP ExAC gnomAD |
|
|
CA405869602 rs761956794 |
101 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA308381827 rs968169579 |
101 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs761956794 CA9441915 |
101 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1186768719 CA405869582 |
103 | I>F | No |
ClinGen TOPMed |
|
|
rs1599985751 CA405869579 |
103 | I>T | No |
ClinGen Ensembl |
|
|
CA405869567 rs1260732312 |
104 | Q>R | No |
ClinGen TOPMed |
|
|
CA9441914 rs762920486 |
105 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs761598534 CA9441911 |
107 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1599985609 CA405869522 |
108 | N>T | No |
ClinGen Ensembl |
|
|
CA405869509 rs1231123715 |
109 | S>N | No |
ClinGen gnomAD |
|
|
CA9441910 rs776161532 |
113 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA308381804 rs1054914884 |
113 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs1257065618 CA405869451 |
114 | A>S | No |
ClinGen gnomAD |
|
|
CA405869454 rs1257065618 |
114 | A>T | No |
ClinGen gnomAD |
|
|
CA308381793 rs937810724 |
114 | A>V | No |
ClinGen TOPMed |
|
|
rs1203244050 CA405869436 |
115 | P>L | No |
ClinGen gnomAD |
|
|
rs141209878 CA9441909 |
116 | G>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs148339167 CA9441908 |
117 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9441906 rs771687909 |
118 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs779749219 CA9441907 |
118 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA9441904 rs778304096 |
119 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA9441905 rs749837210 |
119 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9441902 rs753052094 |
120 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA405869381 rs1390462628 |
120 | M>R | No |
ClinGen gnomAD |
|
|
CA405869374 rs1284479566 |
121 | D>N | No |
ClinGen TOPMed |
|
|
rs201717577 CA9441900 |
122 | Y>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9441901 rs781400894 |
122 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA405869336 rs1473114374 |
124 | C>R | No |
ClinGen gnomAD |
|
|
CA9441899 rs751752149 |
126 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1217996479 CA405869300 |
127 | P>L | No |
ClinGen TOPMed |
|
|
CA308381669 rs908489075 |
128 | S>R | No |
ClinGen Ensembl |
|
|
rs1460139068 CA405869288 |
129 | D>G | No |
ClinGen gnomAD |
|
|
rs984104264 CA308381668 |
130 | S>C | No |
ClinGen TOPMed |
|
|
rs1209358799 CA405869269 |
131 | S>F | No |
ClinGen gnomAD |
|
|
CA9441898 rs768885006 |
132 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9441897 rs143335817 |
132 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA308381646 rs1028818887 |
134 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
CA9441895 rs765159398 |
134 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1028818887 CA405869237 |
134 | E>V | No |
ClinGen TOPMed gnomAD |
|
|
rs761642183 CA9441894 |
136 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs776555398 CA9441893 |
138 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768539227 CA9441892 COSM189287 |
139 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1158640193 CA405869165 |
140 | V>I | No |
ClinGen TOPMed |
|
|
CA308381614 rs867449500 |
144 | R>Q | No |
ClinGen Ensembl |
|
|
CA9441889 rs575460456 COSM996625 |
144 | R>W | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA308381604 rs879199809 |
146 | K>T | No |
ClinGen Ensembl |
|
|
CA405869078 rs1477451871 |
147 | V>A | No |
ClinGen gnomAD |
|
|
CA308381211 rs886949760 |
149 | M>T | No |
ClinGen Ensembl |
|
|
rs1409596764 CA405868549 |
151 | D>N | No |
ClinGen gnomAD |
|
|
rs1599981496 CA405868446 |
156 | K>N | No |
ClinGen Ensembl |
|
|
CA9441866 rs777080235 |
156 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9441864 rs747313418 |
168 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA405868318 rs1305342088 |
169 | V>M | No |
ClinGen gnomAD |
|
|
CA9441862 rs572670425 COSM996623 |
170 | R>W | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs1599981195 CA405868264 |
174 | T>P | No |
ClinGen Ensembl |
|
|
CA405868252 rs1280863854 |
175 | G>D | No |
ClinGen gnomAD |
|
|
rs398123625 COSM1645332 CA221735 |
175 | G>S | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA9441859 rs757233968 |
176 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764058037 CA9441857 |
176 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764058037 CA9441858 |
176 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405868222 rs755987647 |
178 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs991833403 CA308381157 |
179 | A>T | No |
ClinGen TOPMed |
|
|
CA308381155 rs959871626 |
180 | M>V | No |
ClinGen TOPMed |
|
|
rs1352944542 CA405868172 |
183 | L>Q | No |
ClinGen gnomAD |
|
|
rs1470454115 CA405868165 |
184 | R>Q | No |
ClinGen gnomAD |
|
|
rs752597471 CA9441855 |
184 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs55859611 CA308381142 VAR_040356 |
188 | I>V | No |
ClinGen UniProt TOPMed dbSNP gnomAD |
|
|
CA9441853 rs759277032 |
191 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774646421 CA9441805 |
192 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA405868010 rs1225273209 |
192 | D>N | No |
ClinGen gnomAD |
|
|
rs763319755 CA9441803 COSM3422825 |
195 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA405867975 rs1568526119 |
197 | T>A | No |
ClinGen Ensembl |
|
|
rs371943261 CA9441802 |
198 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
CA9441800 rs748177849 |
202 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA9441798 rs549648922 |
207 | T>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs35817154 VAR_040357 CA9441797 |
208 | R>K | No |
ClinGen UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1599969539 CA405867889 |
210 | P>R | No |
ClinGen Ensembl |
|
|
rs1175170174 CA405867875 |
212 | L>P | No |
ClinGen gnomAD |
|
|
rs758767505 CA9441736 |
214 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758767505 CA9441737 |
214 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1189807846 CA405867850 |
215 | L>V | No |
ClinGen gnomAD |
|
|
rs1599963715 CA405867808 |
221 | T>P | No |
ClinGen Ensembl |
|
|
CA9441732 rs769554060 |
222 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs1599963532 CA405867793 |
223 | D>A | No |
ClinGen Ensembl |
|
|
CA9441729 rs545889598 |
223 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs780894376 CA9441730 |
223 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1322643388 CA405867789 |
224 | R>C | No |
ClinGen gnomAD |
|
|
CA405867786 rs746407532 |
224 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA9441728 rs746407532 |
224 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA405867774 rs1373751808 |
226 | C>Y | No |
ClinGen gnomAD |
|
|
rs754286414 CA9441725 |
231 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs1227145174 CA405867726 |
233 | N>D | No |
ClinGen gnomAD |
|
|
rs988297708 CA308378294 |
233 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1363221582 CA405867219 |
243 | R>Q | No |
ClinGen gnomAD |
|
|
rs763896696 CA9441694 |
243 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA9441693 rs760399082 |
245 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs775105991 CA9441692 |
245 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405867173 rs1367814764 |
248 | T>A | No |
ClinGen gnomAD |
|
|
CA9441691 rs771461640 |
248 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA308377603 rs1044041105 |
250 | E>* | No |
ClinGen TOPMed |
|
|
CA308377581 rs751522812 |
251 | R>Q | No |
ClinGen TOPMed |
|
|
CA9441689 rs773553776 |
251 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1428621686 CA405867132 |
252 | A>G | No |
ClinGen gnomAD |
|
|
CA9441688 rs770151213 |
252 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA405867128 rs781527200 |
253 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA9441685 rs145907048 |
253 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781527200 CA9441686 |
253 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1207387979 CA405867059 |
259 | I>V | No |
ClinGen gnomAD |
|
|
CA9441684 rs747207035 |
261 | S>L | No |
ClinGen ExAC |
|
|
CA9441682 rs758367800 |
262 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA308377549 rs1804324 |
265 | Y>N | No |
ClinGen Ensembl |
|
|
CA405866958 rs757245351 |
268 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs757245351 CA9441679 |
268 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA308377525 rs761347839 |
269 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA9441677 rs763951605 |
269 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1347953319 CA405866933 |
271 | V>M | No |
ClinGen gnomAD |
|
|
rs1304063423 CA405866923 |
272 | V>I | No |
ClinGen gnomAD |
|
|
CA9441675 rs774959037 |
274 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1240210533 CA405866828 |
281 | L>V | No |
ClinGen gnomAD |
|
|
CA405866809 rs1473345359 |
284 | D>N | No |
ClinGen TOPMed |
|
|
CA9441636 rs369596328 |
290 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9441635 rs762379584 |
291 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA405866699 rs1333003888 |
299 | E>* | No |
ClinGen gnomAD |
|
|
CA9441634 rs376326333 |
305 | A>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA9441633 rs777865319 |
306 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1378308963 CA405866648 |
307 | M>V | No |
ClinGen gnomAD |
|
|
rs1243812798 CA405866597 |
314 | P>A | No |
ClinGen gnomAD |
|
|
rs140987550 CA308376544 |
314 | P>L | No |
ClinGen ESP TOPMed |
|
|
CA405866589 rs1441119595 |
315 | E>G | No |
ClinGen gnomAD |
|
|
CA405866567 rs1206035617 |
318 | A>V | No |
ClinGen gnomAD |
|
|
rs374756725 CA308376460 |
325 | N>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA308376457 rs980537301 |
326 | D>Y | No |
ClinGen Ensembl |
|
|
rs144395843 CA9441610 |
329 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs763266151 CA9441609 |
330 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1323113005 CA405866456 COSM1481073 |
333 | W>* | breast [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1323113005 CA405866455 |
333 | W>C | No |
ClinGen gnomAD |
|
|
CA405866443 rs1568519097 |
335 | G>R | No |
ClinGen Ensembl |
|
|
rs1004075798 CA308376430 |
340 | M>I | No |
ClinGen Ensembl |
|
|
rs776561628 CA9441605 |
340 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA9441603 rs372625056 |
344 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1166756826 CA405866379 |
344 | M>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1422114933 COSM996621 CA405866381 |
344 | M>V | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1169786300 CA405866366 |
346 | G>S | No |
ClinGen TOPMed |
|
|
rs145305228 CA9441601 |
347 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs202125073 CA308376418 |
347 | R>H | No |
ClinGen 1000Genomes |
|
|
rs745497338 CA9441600 |
349 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1020231672 COSM1579112 CA308376415 |
351 | Y>C | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA9441598 rs756750912 |
352 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA308376411 rs886899893 |
352 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA9441597 rs753149827 |
353 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA405866323 rs1195945663 |
353 | Q>R | No |
ClinGen gnomAD |
|
|
COSM439520 CA405866302 rs1568518786 |
356 | E>K | breast [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1026908617 CA308376403 |
357 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA9441594 rs752039318 |
357 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200433217 CA9441591 |
360 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs930976777 CA308376396 |
360 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA405866251 rs1568518681 |
364 | M>V | No |
ClinGen Ensembl |
|
|
CA9441590 rs765232271 |
366 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA405866226 rs1470923617 |
367 | I>V | No |
ClinGen gnomAD |
|
|
rs144444132 COSM268637 CA9441588 |
368 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC gnomAD |
|
CA9441587 rs768703735 COSM996619 |
368 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA405866205 rs1204214325 |
370 | P>L | No |
ClinGen gnomAD |
|
|
CA9441583 rs745550650 |
371 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA9441582 rs778561687 COSM318640 |
371 | R>L | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs139506765 CA9441581 |
372 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs777146223 CA9441579 |
373 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA308376337 rs200395003 |
376 | E>D | No |
ClinGen Ensembl |
|
|
rs1466949645 CA405866175 |
376 | E>K | No |
ClinGen gnomAD |
|
|
CA9441575 rs758765375 |
377 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs765549797 CA9441573 |
378 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs529742537 CA405866160 |
378 | K>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs529742537 CA9441572 |
378 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs753989242 CA9441571 |
382 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1437512007 CA405866121 |
385 | L>H | No |
ClinGen gnomAD |
|
|
CA405866043 rs1458251691 |
394 | G>C | No |
ClinGen gnomAD |
|
|
rs1257580103 CA405866039 |
395 | G>W | No |
ClinGen gnomAD |
|
|
CA9441524 rs765026585 |
396 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405866033 rs765026585 |
396 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1319165364 CA405866014 |
399 | D>N | No |
ClinGen TOPMed |
|
|
rs1256885928 CA405865995 |
401 | K>R | No |
ClinGen gnomAD |
|
|
CA405865980 rs1599949801 |
403 | V>G | No |
ClinGen Ensembl |
|
|
rs767995122 CA9441521 |
404 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA405865977 rs1328944283 |
404 | M>V | No |
ClinGen gnomAD |
|
|
CA405865964 rs1485158704 |
405 | E>A | No |
ClinGen TOPMed |
|
|
CA405865916 rs1198321649 |
412 | I>L | No |
ClinGen TOPMed |
|
|
CA405865906 rs1470884234 |
413 | N>S | No |
ClinGen gnomAD |
|
|
rs749701848 CA9441515 |
415 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA9441511 rs781260194 |
417 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA9441510 rs754942500 |
418 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs1159523716 CA405865869 |
418 | V>G | No |
ClinGen TOPMed |
|
|
rs779909600 CA9441508 |
419 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA308375887 rs1010551997 |
425 | P>H | No |
ClinGen TOPMed |
|
|
CA9441483 rs752215436 |
428 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA405865783 rs767039609 |
429 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405865784 rs767039609 |
429 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1399189737 CA405865786 |
429 | Q>R | No |
ClinGen gnomAD |
|
|
CA9441481 rs375739695 |
431 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs867381867 CA308375876 |
432 | S>C | No |
ClinGen Ensembl |
|
|
CA405865675 rs1371711287 |
445 | A>P | No |
ClinGen TOPMed |
|
|
rs1316408057 CA405865642 |
450 | I>V | No |
ClinGen TOPMed |
|
|
CA405865630 rs1207581157 |
452 | P>T | No |
ClinGen gnomAD |
|
|
CA405865622 rs1278736092 |
453 | P>S | No |
ClinGen gnomAD |
|
|
rs1278736092 CA405865624 |
453 | P>T | No |
ClinGen gnomAD |
|
|
rs757239082 CA405865609 |
455 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757239082 CA405865610 |
455 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9441469 rs778976738 |
455 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778976738 CA308375838 |
455 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9441470 rs757239082 |
455 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405865184 rs1468068048 |
457 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA9441422 rs756547812 |
458 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs753179882 CA405865173 |
458 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1173481204 CA405865151 |
462 | L>P | No |
ClinGen TOPMed |
|
|
CA405865149 rs1251045508 |
463 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA405865148 rs1251045508 |
463 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA405865121 rs1413205437 |
466 | Q>H | No |
ClinGen TOPMed |
|
|
rs759851333 CA9441419 |
467 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9441420 rs142926499 |
467 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1385936959 CA405865111 |
469 | H>Y | No |
ClinGen gnomAD |
|
|
CA405865101 rs1368133745 |
470 | F>I | No |
ClinGen gnomAD |
|
|
CA405865063 rs1447335111 |
475 | Y>C | No |
ClinGen gnomAD |
|
|
CA405865062 rs1447335111 |
475 | Y>S | No |
ClinGen gnomAD |
|
|
rs766596535 CA9441417 |
476 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA405865044 rs1297243797 |
478 | S>N | No |
ClinGen TOPMed |
|
|
CA9441414 rs769806554 |
480 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405865032 rs769806554 |
480 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA308375219 rs1017033490 |
480 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA308375208 rs1044344132 |
481 | E>K | No |
ClinGen Ensembl |
3 associated diseases with P31751
[MIM: 125853]: Diabetes mellitus, non-insulin-dependent (NIDDM)
A multifactorial disorder of glucose homeostasis caused by a lack of sensitivity to the body's own insulin. Affected individuals usually have an obese body habitus and manifestations of a metabolic syndrome characterized by diabetes, insulin resistance, hypertension and hypertriglyceridemia. The disease results in long-term complications that affect the eyes, kidneys, nerves, and blood vessels. {ECO:0000269|PubMed:15166380, ECO:0000269|PubMed:19164855}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 240900]: Hypoinsulinemic hypoglycemia with hemihypertrophy (HIHGHH)
A disorder characterized by hypoglycemia, low insulin levels, low serum levels of ketone bodies and branched-chain amino acids, left-sided hemihypertrophy, neonatal macrosomia, reduced consciousness and hypoglycemic seizures. {ECO:0000269|PubMed:21979934}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A multifactorial disorder of glucose homeostasis caused by a lack of sensitivity to the body's own insulin. Affected individuals usually have an obese body habitus and manifestations of a metabolic syndrome characterized by diabetes, insulin resistance, hypertension and hypertriglyceridemia. The disease results in long-term complications that affect the eyes, kidneys, nerves, and blood vessels. {ECO:0000269|PubMed:15166380, ECO:0000269|PubMed:19164855}. Note=The disease is caused by variants affecting the gene represented in this entry.
- A disorder characterized by hypoglycemia, low insulin levels, low serum levels of ketone bodies and branched-chain amino acids, left-sided hemihypertrophy, neonatal macrosomia, reduced consciousness and hypoglycemic seizures. {ECO:0000269|PubMed:21979934}. Note=The disease is caused by variants affecting the gene represented in this entry.
7 regional properties for P31751
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 152 - 409 | IPR000719 |
| domain | AGC-kinase, C-terminal | 410 - 481 | IPR000961 |
| active_site | Serine/threonine-protein kinase, active site | 271 - 283 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 158 - 191 | IPR017441 |
| domain | Protein kinase, C-terminal | 430 - 475 | IPR017892 |
| domain | Protein Kinase B beta, catalytic domain | 156 - 478 | IPR034677 |
| domain | Protein Kinase B, pleckstrin homology domain | 4 - 111 | IPR039026 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24351 | RIBOSOMAL PROTEIN S6 KINASE |
| PANTHER Subfamily | PTHR24351:SF192 | PROTEIN KINASE C |
| PANTHER Protein Class | protein modifying enzyme | |
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
32 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| carbohydrate transport | The directed movement of carbohydrate into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. Carbohydrates are a group of organic compounds based of the general formula Cx(H2O)y. |
| cellular response to high light intensity | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a high light intensity stimulus. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| fat cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of an adipocyte, an animal connective tissue cell specialized for the synthesis and storage of fat. |
| glucose metabolic process | The chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. D-glucose is dextrorotatory and is sometimes known as dextrose; it is an important source of energy for living organisms and is found free as well as combined in homo- and hetero-oligosaccharides and polysaccharides. |
| glycogen biosynthetic process | The chemical reactions and pathways resulting in the formation of glycogen, a polydisperse, highly branched glucan composed of chains of D-glucose residues. |
| insulin receptor signaling pathway | The series of molecular signals generated as a consequence of the insulin receptor binding to insulin. |
| intracellular protein transmembrane transport | The directed movement of proteins in a cell, from one side of a membrane to another by means of some agent such as a transporter or pore. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mammary gland epithelial cell differentiation | The process in which a relatively unspecialized epithelial cell becomes a more specialized epithelial cell of the mammary gland. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of long-chain fatty acid import across plasma membrane | Any process that decreases the rate, frequency or extent of plasma membrane long-chain fatty acid transport. Plasma membrane long-chain fatty acid transport is the directed movement of long-chain fatty acids across the plasma membrane. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peripheral nervous system myelin maintenance | The process in which the structure and material content of mature peripheral nervous system myelin is kept in a functional state. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell motility | Any process that activates or increases the frequency, rate or extent of cell motility. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of fatty acid beta-oxidation | Any process that activates or increases the frequency, rate or extent of fatty acid beta-oxidation. |
| positive regulation of glucose import | Any process that activates or increases the frequency, rate or extent of the import of the hexose monosaccharide glucose into a cell or organelle. |
| positive regulation of glucose metabolic process | Any process that increases the rate, frequency or extent of glucose metabolism. Glucose metabolic processes are the chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. |
| positive regulation of glycogen biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of glycogen. |
| positive regulation of mitochondrial membrane potential | Any process that activates or increases the frequency, rate or extent of establishment or extent of a mitochondrial membrane potential, the electric potential existing across any mitochondrial membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| positive regulation of protein targeting to membrane | Any process that increases the frequency, rate or extent of the process of directing proteins towards a membrane, usually using signals contained within the protein. |
| positive regulation of vesicle fusion | Any process that activates or increases the frequency, rate or extent of vesicle fusion. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
| regulation of translation | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| retinal rod cell apoptotic process | Any apoptotic process in a retinal rod cell, one of the two photoreceptor cell types of the vertebrate retina. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
32 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q01314 | AKT1 | RAC-alpha serine/threonine-protein kinase | Bos taurus (Bovine) | SS |
| Q8INB9 | Akt | RAC serine/threonine-protein kinase | Drosophila melanogaster (Fruit fly) | SS |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens (Human) | EV |
| Q05513 | PRKCZ | Protein kinase C zeta type | Homo sapiens (Human) | SS |
| P41743 | PRKCI | Protein kinase C iota type | Homo sapiens (Human) | EV |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| Q96BR1 | SGK3 | Serine/threonine-protein kinase Sgk3 | Homo sapiens (Human) | SS |
| Q9HBY8 | SGK2 | Serine/threonine-protein kinase Sgk2 | Homo sapiens (Human) | SS |
| O00141 | SGK1 | Serine/threonine-protein kinase Sgk1 | Homo sapiens (Human) | PR |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | Homo sapiens (Human) | EV |
| Q9Y2H1 | STK38L | Serine/threonine-protein kinase 38-like | Homo sapiens (Human) | EV |
| Q6A1A2 | PDPK2P | Putative 3-phosphoinositide-dependent protein kinase 2 | Homo sapiens (Human) | PR |
| P31750 | Akt1 | RAC-alpha serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9WUA6 | Akt3 | RAC-gamma serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q60823 | Akt2 | RAC-beta serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| P47196 | Akt1 | RAC-alpha serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| Q63484 | Akt3 | RAC-gamma serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| P47197 | Akt2 | RAC-beta serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| Q9XTG7 | akt-2 | Serine/threonine-protein kinase akt-2 | Caenorhabditis elegans | SS |
| Q17941 | akt-1 | Serine/threonine-protein kinase akt-1 | Caenorhabditis elegans | PR |
| Q9SUA3 | D6PKL1 | Serine/threonine-protein kinase D6PKL1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNEVSVIKEG | WLHKRGEYIK | TWRPRYFLLK | SDGSFIGYKE | RPEAPDQTLP | PLNNFSVAEC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QLMKTERPRP | NTFVIRCLQW | TTVIERTFHV | DSPDEREEWM | RAIQMVANSL | KQRAPGEDPM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DYKCGSPSDS | STTEEMEVAV | SKARAKVTMN | DFDYLKLLGK | GTFGKVILVR | EKATGRYYAM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KILRKEVIIA | KDEVAHTVTE | SRVLQNTRHP | FLTALKYAFQ | THDRLCFVME | YANGGELFFH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LSRERVFTEE | RARFYGAEIV | SALEYLHSRD | VVYRDIKLEN | LMLDKDGHIK | ITDFGLCKEG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ISDGATMKTF | CGTPEYLAPE | VLEDNDYGRA | VDWWGLGVVM | YEMMCGRLPF | YNQDHERLFE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LILMEEIRFP | RTLSPEAKSL | LAGLLKKDPK | QRLGGGPSDA | KEVMEHRFFL | SINWQDVVQK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KLLPPFKPQV | TSEVDTRYFD | DEFTAQSITI | TPPDRYDSLG | LLELDQRTHF | PQFSYSASIR |
| E |