P31380
Gene name |
FUN30 (YAL019W, YAL001) |
Protein name |
ATP-dependent helicase FUN30 |
Names |
|
Species |
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) |
KEGG Pathway |
sce:YAL019W |
EC number |
3.6.4.12: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
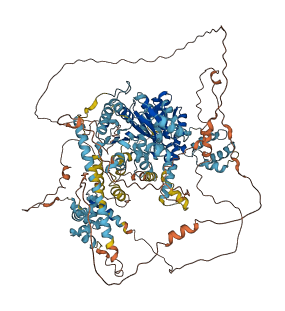
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P31380
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 5GN1 | X-ray | 195 A | A/B/C/D | 780-1122 | PDB |
| AF-P31380-F1 | Predicted | AlphaFoldDB |
20 variants for P31380
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| s01-114929 | 4 | S>P | No | SGRP | |
| s01-115110 | 64 | K>T | No | SGRP | |
| s01-115591 | 224 | E>D | No | SGRP | |
| s01-115907 | 330 | D>N | No | SGRP | |
| s01-115918 | 333 | E>D | No | SGRP | |
| s01-115922 | 335 | A>T | No | SGRP | |
| s01-116176 | 419 | N>K | No | SGRP | |
| s01-116179 | 420 | N>K | No | SGRP | |
| s01-116202 | 428 | A>G | No | SGRP | |
| s01-116207 | 430 | A>T | No | SGRP | |
| s01-116328 | 470 | I>T | No | SGRP | |
| s01-116458 | 513 | N>K | No | SGRP | |
| s01-116521 | 534 | F>L | No | SGRP | |
| s01-116549 | 544 | I>V | No | SGRP | |
| s01-116601 | 561 | G>D | No | SGRP | |
| s01-116778 | 620 | P>L | No | SGRP | |
| s01-116906 | 663 | D>H | No | SGRP | |
| s01-117255 | 779 | N>T | No | SGRP | |
| s01-117500 | 861 | S>P | No | SGRP | |
| s01-118305 | 1129 | T>I | No | SGRP |
No associated diseases with P31380
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.12 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromosome, centromeric region | The region of a chromosome that includes the centromeric DNA and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome. |
| chromosome, telomeric region | The end of a linear chromosome, required for the integrity and maintenance of the end. A chromosome telomere usually includes a region of telomerase-encoded repeats the length of which rarely exceeds 20 bp each and that permits the formation of a telomeric loop (T-loop). The telomeric repeat region is usually preceded by a sub-telomeric region that is gene-poor but rich in repetitive elements. Some telomeres only consist of the latter part (for eg. D. melanogaster telomeres). |
| mating-type region heterochromatin | Heterochromatic regions of the chromosome found at silenced mating-type loci. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| ATP-dependent activity, acting on DNA | Catalytic activity that acts to modify DNA, driven by ATP hydrolysis. |
| ATP-dependent chromatin remodeler activity | An activity, driven by ATP hydrolysis, that modulates the contacts between histones and DNA, resulting in a change in chromosome architecture within the nucleosomal array, leading to chromatin remodeling. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA helicase activity | Unwinding of a DNA helix, driven by ATP hydrolysis. |
| identical protein binding | Binding to an identical protein or proteins. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| chromatin remodeling | A dynamic process of chromatin reorganization resulting in changes to chromatin structure. These changes allow DNA metabolic processes such as transcriptional regulation, DNA recombination, DNA repair, and DNA replication. |
| DNA double-strand break processing | The 5' to 3' exonucleolytic resection of the DNA at the site of the break to form a 3' single-strand DNA overhang. |
| heterochromatin assembly | An epigenetic gene silencing mechanism in which chromatin is compacted into heterochromatin, resulting in a chromatin conformation refractory to transcription. This process starts with heterochromatin nucleation, its spreading, and ends with heterochromatin boundary formation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of RNA splicing | Any process that activates or increases the frequency, rate or extent of RNA splicing. |
| rDNA heterochromatin assembly | The formation of heterochromatin at ribosomal DNA, characterized by the modified histone H3K9me3. |
| silent mating-type cassette heterochromatin assembly | Repression of transcription at silent mating-type loci by alteration of the structure of chromatin. |
| subtelomeric heterochromatin assembly | The compaction of chromatin into heterochromatin at the subtelomeric region. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32597 | STH1 | Nuclear protein STH1/NPS1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P38144 | ISW1 | ISWI chromatin-remodeling complex ATPase ISW1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q9VL72 | Etl1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 homolog | Drosophila melanogaster (Fruit fly) | PR |
| Q9H4L7 | SMARCAD1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 | Homo sapiens (Human) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGSHSNDED | DVVQVPETSS | PTKVASSSPL | KPTSPTVPDA | SVASLRSRFT | FKPSDPSEGA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| HTSKPLPSGS | PEVALVNLAR | EFPDFSQTLV | QAVFKSNSFN | LQSARERLTR | LRQQRQNWTW |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NKNASPKKSE | TPPPVKKSLP | LANTGRLSSI | HGNINNKSSK | ITVAKQKTSI | FDRYSNVINQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KQYTFELPTN | LNIDSEALSK | LPVNYNKKRR | LVRADQHPIG | KSYESSATQL | GSAREKLLAN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RKYGRHANDN | DEEEEESMMT | DDDDASGDDY | TESTPQINLD | EQVLQFINDS | DIVDLSDLSD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TTMHKAQLIA | SHRPYSSLNA | FVNTNFNDKD | TEENASNKRK | RRAAASANES | ERLLDKITQS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IRGYNAIESV | IKKCSSYGDL | VTSQMKKWGV | QVEGDNSELD | LMNLGEDDDD | DNDDGNNDNN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NSNNNNTAGA | DATSKEKEDT | KAVVEGFDET | SAEPTPAPAP | APVERETKRI | RNTTKPKVVE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DEDDDVDLEA | IDDELPQSEH | EDDDYEEEDE | DYNDEEEDVE | YDDGDDDDDD | DDEFVATRKN |
| 550 | 560 | 570 | 580 | 590 | 600 |
| THVISTTSRN | GRKPIVKFFK | GKPRLLSPEI | SLKDYQQTGI | NWLNLLYQNK | MSCILADDMG |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LGKTCQVISF | FAYLKQINEP | GPHLVVVPSS | TLENWLREFQ | KFAPALKIEP | YYGSLQEREE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LRDILERNAG | KYDVIVTTYN | LAAGNKYDVS | FLKNRNFNVV | VYDEGHMLKN | STSERFAKLM |
| 730 | 740 | 750 | 760 | 770 | 780 |
| KIRANFRLLL | TGTPLQNNLK | ELMSLLEFIM | PNLFISKKES | FDAIFKQRAK | TTDDNKNHNP |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LLAQEAITRA | KTMMKPFILR | RRKDQVLKHL | PPKHTHIQYC | ELNAIQKKIY | DKEIQIVLEH |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KRMIKDGELP | KDAKEKSKLQ | SSSSKNLIMA | LRKASLHPLL | FRNIYNDKII | TKMSDAILDE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| PAYAENGNKE | YIKEDMSYMT | DFELHKLCCN | FPNTLSKYQL | HNDEWMQSGK | IDALKKLLKT |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| IIVDKQEKVL | IFSLFTQVLD | ILEMVLSTLD | YKFLRLDGST | QVNDRQLLID | KFYEDKDIPI |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| FILSTKAGGF | GINLVCANNV | IIFDQSFNPH | DDRQAADRAH | RVGQTKEVNI | TTLITKDSIE |
| 1090 | 1100 | 1110 | 1120 | 1130 | |
| EKIHQLAKNK | LALDSYISED | KKSQDVLESK | VSDMLEDIIY | DENSKPKGTK | E |