P31268
Gene name |
HOXA7 (HOX1A) |
Protein name |
Homeobox protein Hox-A7 |
Names |
Homeobox protein Hox 1.1, Homeobox protein Hox-1A |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:3204 |
EC number |
|
Protein Class |
|
Descriptions
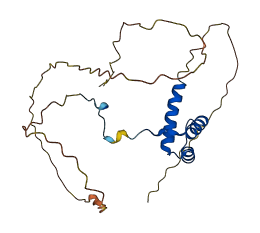
HoxA7 is a sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. This protein possesses intrinsically disordered regions (IDRs) with large negative charge, some of which involve a consecutive sequence of aspartate (D) or glutamate (E) residues, known as D/E repeats. These D/E repeats can cause autoinhibition through intramolecular electrostatic interaction with HMG boxes and modulate binding to DNA. This autoinhibited state can transition into the uninhibited complex with DNA through an electrostatically driven induced-fit process, which accelerates the target DNA search kinetics in the presence of non-functional high-affinity ligands ('decoys').
Autoinhibitory domains (AIDs)
Target domain |
128-192 (DNA-binding domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for P31268
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P31268-F1 | Predicted | AlphaFoldDB |
276 variants for P31268
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs149657061 | 2 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs781611799 | 2 | S>I | No |
ExAC gnomAD |
|
| rs781611799 | 2 | S>N | No |
ExAC gnomAD |
|
| rs757369844 | 3 | S>Y | No |
ExAC gnomAD |
|
| rs866752726 | 4 | S>L | No |
TOPMed gnomAD |
|
| rs751666779 | 4 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs758495730 | 8 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs765702602 | 9 | A>G | No |
ExAC gnomAD |
|
| rs907646153 | 9 | A>T | No | TOPMed | |
| rs1309115771 | 10 | L>R | No |
TOPMed gnomAD |
|
| rs759944196 | 12 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs371532380 | 13 | K>R | No |
ESP ExAC gnomAD |
|
| rs377689779 | 14 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1161572722 | 15 | T>A | No | gnomAD | |
| rs1379798108 | 16 | A>G | No | gnomAD | |
| rs145778968 | 17 | G>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145778968 | 17 | G>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs78410337 | 17 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs78410337 | 17 | G>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2301721 | 18 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
VAR_028001 rs2301721 |
18 | A>T | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs550774120 | 19 | S>A | No |
1000Genomes gnomAD |
|
| rs745678681 | 21 | F>I | No |
ExAC gnomAD |
|
| rs530671173 | 24 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1284749326 | 25 | E>* | No | gnomAD | |
| rs771200839 | 25 | E>G | No |
ExAC gnomAD |
|
| rs1284749326 | 25 | E>Q | No | gnomAD | |
| rs747255380 | 26 | P>S | No |
ExAC gnomAD |
|
| rs758448332 | 27 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs758448332 | 27 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs979099678 | 29 | C>F | No | TOPMed | |
| rs752752010 | 29 | C>G | No |
ExAC gnomAD |
|
|
COSM4824345 rs755438662 |
30 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs866074728 | 32 | A>V | No | Ensembl | |
| rs761087427 | 33 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs766593338 | 33 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs193124013 | 34 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1018055387 | 36 | Q>R | No | TOPMed | |
| rs1422982625 | 38 | S>R | No |
TOPMed gnomAD |
|
| rs765294385 | 39 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs765294385 | 39 | G>R | No |
ExAC gnomAD |
|
| rs765294385 | 39 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs139653572 | 39 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs770701724 | 40 | Y>* | No |
ExAC gnomAD |
|
| rs776620721 | 40 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs776620721 | 40 | Y>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 41 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746792177 | 41 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs746792177 | 41 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs748260443 | 42 | A>V | No |
ExAC gnomAD |
|
| rs1346956477 | 43 | G>D | No | gnomAD | |
| COSM3928998 | 44 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778899843 | 44 | A>V | No |
ExAC gnomAD |
|
| rs1207033824 | 45 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| COSM6110128 | 46 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1233857798 | 46 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs879105208 | 47 | F>V | No | Ensembl | |
| rs756474411 | 48 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs749850344 | 48 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs749850344 | 48 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs756474411 | 48 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs878972024 | 49 | S>A | No | Ensembl | |
| rs750774350 | 49 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs1029466417 | 51 | V>A | No | Ensembl | |
| rs764900202 | 52 | P>Q | No |
ExAC gnomAD |
|
| rs1457386778 | 53 | G>S | No | gnomAD | |
| rs759115973 | 53 | G>V | No | ExAC | |
| rs1172350298 | 55 | Y>C | No | gnomAD | |
| rs776477605 | 55 | Y>H | No |
ExAC gnomAD |
|
| rs766322170 | 56 | N>K | No |
ExAC gnomAD |
|
| rs1476627731 | 56 | N>S | No |
TOPMed gnomAD |
|
| TCGA novel | 57 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760410079 | 57 | V>G | No |
ExAC gnomAD |
|
| COSM1088874 | 57 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772288938 | 58 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs1269033672 | 58 | N>S | No | gnomAD | |
| COSM1312998 | 58 | N>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1209746860 | 59 | S>N | No | gnomAD | |
| rs748309582 | 59 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1562738126 | 60 | P>T | No | Ensembl | |
| rs1253448405 | 61 | L>F | No | gnomAD | |
| rs1344928638 | 63 | Q>H | No | Ensembl | |
| rs559354439 | 64 | S>N | No |
1000Genomes ExAC gnomAD |
|
| rs768791120 | 64 | S>R | No |
ExAC TOPMed gnomAD |
|
| COSM1088873 | 65 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM600783 COSM6110130 rs1384986017 |
65 | P>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1038275564 | 67 | A>V | No | Ensembl | |
| rs756592104 | 69 | G>C | No |
ExAC gnomAD |
|
| rs1006250755 | 69 | G>V | No | TOPMed | |
| rs1359389813 | 70 | Y>N | No | TOPMed | |
| rs781551044 | 71 | G>D | No |
ExAC gnomAD |
|
| TCGA novel | 72 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1274092644 | 74 | A>G | No | TOPMed | |
| rs752468389 | 74 | A>T | No |
ExAC gnomAD |
|
| rs1583421468 | 75 | D>A | No | Ensembl | |
| COSM746308 | 75 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1399312973 | 76 | A>T | No | gnomAD | |
| rs529254085 | 76 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs563708052 | 77 | Y>* | No |
ExAC gnomAD |
|
|
rs766259696 COSM1088872 |
78 | G>D | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs753340327 | 78 | G>S | No |
ExAC gnomAD |
|
| rs760595034 | 79 | N>D | No | ExAC | |
| TCGA novel | 79 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1420195665 | 79 | N>I | No |
TOPMed gnomAD |
|
| rs1420195665 | 79 | N>S | No |
TOPMed gnomAD |
|
|
rs750200293 COSM1088871 |
82 | C>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs750200293 | 82 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs1434462964 | 83 | A>T | No | gnomAD | |
| rs921688382 | 83 | A>V | No | TOPMed | |
| rs144742215 | 84 | S>A | No |
ESP gnomAD |
|
| rs144742215 | 84 | S>T | No |
ESP gnomAD |
|
| rs974835724 | 85 | Y>* | No | TOPMed | |
| rs1450272157 | 86 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs767355645 | 86 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1469095750 | 87 | Q>R | No | TOPMed | |
| rs761589955 | 89 | I>M | No |
ExAC gnomAD |
|
| TCGA novel | 89 | I>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1045191807 | 90 | P>A | No | gnomAD | |
| rs1239787579 | 90 | P>L | No |
TOPMed gnomAD |
|
| rs1045191807 | 90 | P>S | No | gnomAD | |
| COSM400291 | 91 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1311317501 | 91 | G>W | No | gnomAD | |
| rs762960362 | 93 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs762960362 | 93 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1056530813 | 94 | S>G | No | gnomAD | |
| rs1337197232 | 95 | D>A | No | gnomAD | |
| rs1171656723 | 99 | G>D | No | gnomAD | |
| rs1342947011 | 99 | G>S | No | TOPMed | |
| rs746326146 | 100 | A>T | No |
ExAC gnomAD |
|
| rs781764041 | 100 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs377596605 | 101 | C>* | No |
ESP ExAC gnomAD |
|
| rs935605212 | 101 | C>Y | No | gnomAD | |
| rs1439661851 | 102 | D>N | No | gnomAD | |
| rs747359929 | 103 | K>E | No |
ExAC gnomAD |
|
|
rs35510017 COSM452920 |
103 | K>R | breast [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs754637278 | 104 | T>M | No |
ExAC gnomAD |
|
| rs769859237 | 105 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1222044457 | 105 | D>N | No | TOPMed | |
| rs750440722 | 106 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs779561079 | 106 | E>Q | No |
ExAC gnomAD |
|
| rs1000360737 | 108 | A>S | No |
TOPMed gnomAD |
|
| rs1000360737 | 108 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 108 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761645013 | 109 | L>P | No |
ExAC gnomAD |
|
| TCGA novel | 110 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1338494214 | 111 | G>V | No | gnomAD | |
| rs769984771 | 112 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1295878557 | 114 | E>A | No | gnomAD | |
| rs1562737958 | 114 | E>Q | No | Ensembl | |
| rs777330616 | 115 | A>T | No |
ExAC gnomAD |
|
| rs771520037 | 115 | A>V | No |
ExAC gnomAD |
|
| rs1471991218 | 117 | F>S | No | gnomAD | |
| rs1196973946 | 118 | R>G | No | TOPMed | |
| rs778216909 | 118 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs778216909 | 118 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1277880163 | 120 | Y>N | No | gnomAD | |
| rs779567620 | 121 | P>H | No | ExAC | |
| rs749030294 | 121 | P>S | No |
ExAC gnomAD |
|
| rs755689640 | 122 | W>R | No |
ExAC gnomAD |
|
| rs1216063088 | 123 | M>L | No | gnomAD | |
| rs544001319 | 124 | R>P | No |
1000Genomes ExAC gnomAD |
|
| rs544001319 | 124 | R>Q | No |
1000Genomes ExAC gnomAD |
|
| rs865788066 | 124 | R>W | No | gnomAD | |
| rs1401927444 | 125 | S>F | No | TOPMed | |
| rs772553123 | 127 | G>A | No | ExAC | |
| TCGA novel | 127 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs900847750 | 128 | P>L | No | Ensembl | |
| rs1036671085 | 129 | D>E | No |
TOPMed gnomAD |
|
| TCGA novel | 130 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs939771927 | 132 | R>Q | No | Ensembl | |
| rs1562737529 | 132 | R>W | No | Ensembl | |
| rs745409883 | 133 | G>D | No |
ExAC gnomAD |
|
| rs780609860 | 134 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1276150189 | 136 | T>P | No | gnomAD | |
| rs982907728 | 137 | Y>C | No | TOPMed | |
| rs1331097182 | 138 | T>M | No |
TOPMed gnomAD |
|
| rs762508295 | 139 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1407134241 | 139 | R>S | No | gnomAD | |
| rs747004706 | 141 | Q>* | No |
ExAC gnomAD |
|
| rs747004706 | 141 | Q>E | No |
ExAC gnomAD |
|
| rs777542472 | 142 | T>A | No |
ExAC gnomAD |
|
| rs1170206862 | 142 | T>M | No | gnomAD | |
| TCGA novel | 146 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs878859385 | 149 | F>Y | No | Ensembl | |
|
rs1477746824 COSM3880409 |
150 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs765531291 | 153 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1179211280 | 153 | R>P | No | gnomAD | |
| rs765531291 | 153 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1583420652 | 154 | Y>S | No | Ensembl | |
| rs753980676 | 155 | L>P | No |
ExAC gnomAD |
|
| rs766627218 | 156 | T>A | No |
ExAC gnomAD |
|
|
COSM3880408 rs559344881 |
156 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs559344881 | 156 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774535792 | 157 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs1343684367 COSM317468 |
157 | R>L | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1343684367 | 157 | R>Q | No |
TOPMed gnomAD |
|
| rs774535792 | 157 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs769491087 | 158 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1449685607 | 158 | R>H | No | gnomAD | |
| rs576810987 | 159 | R>H | No |
1000Genomes ExAC gnomAD |
|
| COSM746309 | 159 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM184301 rs1361050955 |
160 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA gnomAD |
| rs1368746193 | 160 | R>S | No | TOPMed | |
| rs919990927 | 161 | I>T | No | TOPMed | |
|
COSM333707 rs201959803 |
161 | I>V | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs375101630 | 163 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs758229054 | 166 | A>P | No |
ExAC gnomAD |
|
| TCGA novel | 166 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1568450 | 166 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754810820 | 170 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1335089145 | 170 | T>S | No | TOPMed | |
|
rs780389627 COSM4414555 |
172 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1562737385 | 172 | R>H | No | Ensembl | |
| TCGA novel | 174 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750418651 | 175 | K>R | No |
ExAC gnomAD |
|
| COSM1450195 | 176 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768096835 | 177 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs768096835 | 177 | W>C | No |
ExAC TOPMed gnomAD |
|
| rs1309185969 | 179 | Q>H | No | gnomAD | |
| rs751967060 | 181 | R>G | No |
ExAC gnomAD |
|
| COSM1254349 | 181 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1562737367 | 181 | R>L | No | Ensembl | |
|
rs764422900 COSM1088869 |
182 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs764422900 | 182 | R>L | No |
ExAC TOPMed gnomAD |
|
| COSM1088868 | 184 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs927986090 | 184 | K>N | No | TOPMed | |
| COSM1088867 | 186 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1301265808 | 187 | K>T | No | gnomAD | |
| rs374828544 | 189 | H>R | No | ESP | |
| rs776316860 | 190 | K>N | No |
ExAC gnomAD |
|
| rs1378345862 | 191 | D>H | No | gnomAD | |
|
COSM3637739 rs770523270 |
192 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA |
| rs1031543455 | 193 | G>D | No |
TOPMed gnomAD |
|
| rs760210246 | 193 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1031543455 | 193 | G>V | No |
TOPMed gnomAD |
|
| rs571549143 | 194 | P>L | No |
ExAC gnomAD |
|
| rs978085273 | 195 | T>I | No | TOPMed | |
| rs772065138 | 196 | A>G | No |
ExAC TOPMed gnomAD |
|
| COSM1088866 | 196 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs147555041 | 197 | A>S | No |
1000Genomes ExAC gnomAD |
|
| rs749066787 | 200 | P>A | No |
ExAC gnomAD |
|
| rs749066787 | 200 | P>S | No |
ExAC gnomAD |
|
| rs200468253 | 201 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs200468253 | 201 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs371795704 | 203 | A>S | No |
ESP gnomAD |
|
| rs750517268 | 203 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs757346004 | 204 | V>L | No |
ExAC gnomAD |
|
| rs1341434143 | 205 | P>L | No | gnomAD | |
| rs764552031 | 206 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs954358815 | 207 | A>V | No | Ensembl | |
| rs368771286 | 208 | A>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs368771286 | 208 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs138411797 | 209 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1342151391 | 209 | A>V | No | TOPMed | |
| rs1423826860 | 212 | A>G | No | gnomAD | |
| rs1423826860 | 212 | A>V | No | gnomAD | |
| rs370998918 | 213 | A>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs760187331 | 213 | A>S | No |
ExAC gnomAD |
|
| rs760187331 | 213 | A>T | No |
ExAC gnomAD |
|
| rs370998918 | 213 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs771551984 | 214 | D>N | No |
ExAC gnomAD |
|
| rs115619001 | 215 | K>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370786847 | 217 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs140671509 | 217 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs140671509 | 217 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs565645084 | 218 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs565645084 | 218 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs757318981 | 219 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs781471186 | 219 | E>G | No |
ExAC gnomAD |
|
| COSM1241276 | 219 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200815896 | 221 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1265710721 | 221 | D>V | No | TOPMed | |
| rs146964934 | 225 | E>D | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs778284810 | 227 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs143079352 | 228 | E>K | No |
ESP ExAC gnomAD |
|
| rs1562737167 | 229 | E>D | No | Ensembl |
No associated diseases with P31268
No regional properties for P31268
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P31268 | |||
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nuclear membrane | Either of the lipid bilayers that surround the nucleus and form the nuclear envelope; excludes the intermembrane space. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| embryonic skeletal system morphogenesis | The process in which the anatomical structures of the skeleton are generated and organized during the embryonic phase. |
| negative regulation of cell-matrix adhesion | Any process that stops, prevents, or reduces the rate or extent of cell adhesion to the extracellular matrix. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| negative regulation of keratinocyte differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of keratinocyte differentiation. |
| negative regulation of leukocyte migration | Any process that stops, prevents, or reduces the frequency, rate, or extent of leukocyte migration. |
| negative regulation of monocyte differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of monocyte differentiation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| stem cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a stem cell. A stem cell is a cell that retains the ability to divide and proliferate throughout life to provide progenitor cells that can differentiate into specialized cells. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P23459 | HOXD8 | Homeobox protein Hox-D8 | Gallus gallus (Chicken) | PR |
| A2T6Z0 | HOXB1 | Homeobox protein Hox-B1 | Pan troglodytes (Chimpanzee) | SS |
| P10105 | lab | Homeotic protein labial | Drosophila melanogaster (Fruit fly) | EV |
| P49639 | HOXA1 | Homeobox protein Hox-A1 | Homo sapiens (Human) | SS |
| P14653 | HOXB1 | Homeobox protein Hox-B1 | Homo sapiens (Human) | SS |
| P09022 | Hoxa1 | Homeobox protein Hox-A1 | Mus musculus (Mouse) | SS |
| O08656 | Hoxa1 | Homeobox protein Hox-A1 | Rattus norvegicus (Rat) | SS |
| A2D649 | HOXB1 | Homeobox protein Hox-B1 | Macaca mulatta (Rhesus macaque) | SS |
| Q28IU6 | hoxd1 | Homeobox protein Hox-D1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q8JH55 | hoxb8b | Homeobox protein Hox-B8b | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| F1Q4R9 | meox1 | Homeobox protein MOX-1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSSSYYVNAL | FSKYTAGASL | FQNAEPTSCS | FAPNSQRSGY | GAGAGAFAST | VPGLYNVNSP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LYQSPFASGY | GLGADAYGNL | PCASYDQNIP | GLCSDLAKGA | CDKTDEGALH | GAAEANFRIY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PWMRSSGPDR | KRGRQTYTRY | QTLELEKEFH | FNRYLTRRRR | IEIAHALCLT | ERQIKIWFQN |
| 190 | 200 | 210 | 220 | ||
| RRMKWKKEHK | DEGPTAAAAP | EGAVPSAAAT | AAADKADEED | DDEEEEDEEE |