P30679
Gene name |
GNA15 (GNA16) |
Protein name |
Guanine nucleotide-binding protein subunit alpha-15 |
Names |
G alpha-15 , G-protein subunit alpha-15 , Epididymis tissue protein Li 17E , Guanine nucleotide-binding protein subunit alpha-16 , G alpha-16 , G-protein subunit alpha-16 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2769 |
EC number |
|
Protein Class |
|
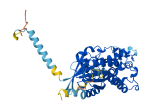
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
192-368 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Goricanec D et al. (2016) "Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding", Proceedings of the National Academy of Sciences of the United States of America, 113, E3629-38
- Coleman DE et al. (1999) "Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex", The Journal of biological chemistry, 274, 16669-72
- Lutz S et al. (2007) "Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs", Science (New York, N.Y.), 318, 1923-7
Autoinhibited structure

Activated structure

1 structures for P30679
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P30679-F1 | Predicted | AlphaFoldDB |
452 variants for P30679
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1360472964 | 3 | R>C | No |
TOPMed gnomAD |
|
| rs75723333 | 3 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs75723333 | 3 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1359117471 | 4 | S>L | No | gnomAD | |
| rs1914460664 | 7 | W>R | No | Ensembl | |
| rs1914460721 | 7 | W>S | No | TOPMed | |
| rs376509314 | 8 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs368219827 | 8 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs376509314 | 8 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1240374665 | 9 | C>* | No | gnomAD | |
| rs1192549861 | 9 | C>R | No | gnomAD | |
| rs909320865 | 11 | P>L | No | TOPMed | |
| rs1474389894 | 12 | W>C | No | gnomAD | |
| rs1472159971 | 12 | W>R | No | gnomAD | |
| rs1159293046 | 12 | W>S | No |
TOPMed gnomAD |
|
| rs758392333 | 15 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs751274515 | 16 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs979316332 | 17 | D>E | No | Ensembl | |
| rs755254026 | 17 | D>V | No |
ExAC gnomAD |
|
| rs1914461534 | 18 | E>A | No |
TOPMed gnomAD |
|
| rs1362058901 | 19 | K>M | No | gnomAD | |
| rs1400840530 | 20 | A>V | No | gnomAD | |
| rs1324102337 | 21 | A>D | No | gnomAD | |
| rs1914461790 | 21 | A>T | No | Ensembl | |
| rs374928184 | 22 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs374928184 | 22 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs771387678 | 23 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs771387678 | 23 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs749833575 | 23 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1599316388 | 24 | V>G | No | Ensembl | |
| rs1351694404 | 24 | V>L | No |
TOPMed gnomAD |
|
| rs1351694404 | 24 | V>M | No |
TOPMed gnomAD |
|
| rs866327080 | 26 | Q>H | No | TOPMed | |
| rs1204709662 | 26 | Q>R | No | gnomAD | |
| rs200281819 | 27 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1444156721 | 28 | I>T | No |
TOPMed gnomAD |
|
| rs1205539525 | 29 | N>K | No |
TOPMed gnomAD |
|
| rs1914462784 | 30 | R>K | No | Ensembl | |
| rs1243645856 | 31 | I>T | No | gnomAD | |
| rs1914462950 | 33 | L>S | No | Ensembl | |
| rs746158062 | 34 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs746158062 | 34 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs768354703 | 34 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1157589821 | 35 | Q>H | No | gnomAD | |
| rs1382167803 | 37 | K>Q | No |
TOPMed gnomAD |
|
| rs369043343 | 38 | Q>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs761540533 | 40 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs761540533 | 40 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs764892056 | 40 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs764892056 | 40 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs764892056 | 40 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1336689193 | 41 | G>E | No | gnomAD | |
| rs766182128 | 41 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs766182128 | 41 | G>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 42 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1382366407 | 44 | K>E | No | Ensembl | |
| rs751384202 | 46 | L>P | No |
ExAC gnomAD |
|
|
rs1224199384 COSM1711603 |
48 | L>F | skin [Cosmic] | No |
cosmic curated gnomAD |
|
TCGA novel rs1347269720 |
48 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| TCGA novel | 50 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2144850916 | 51 | G>D | No | Ensembl | |
| rs772875677 | 52 | E>A | No |
ExAC TOPMed |
|
| rs953694957 | 52 | E>K | No | gnomAD | |
| rs1226527441 | 53 | S>C | No | gnomAD | |
| rs912651393 | 53 | S>R | No | gnomAD | |
|
rs1462808020 COSM1392338 |
54 | G>R | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1361276117 | 54 | G>V | No | gnomAD | |
| rs1222837047 | 56 | S>G | No | gnomAD | |
| rs1290547277 | 56 | S>I | No |
TOPMed gnomAD |
|
| rs1290547277 | 56 | S>N | No |
TOPMed gnomAD |
|
| rs1218721506 | 57 | T>A | No |
TOPMed gnomAD |
|
| rs1489677427 | 59 | I>V | No | gnomAD | |
| rs1914791565 | 60 | K>R | No | gnomAD | |
| rs866353382 | 61 | Q>K | No | Ensembl | |
| rs762498774 | 61 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1428444766 | 62 | M>I | No |
TOPMed gnomAD |
|
| rs1914792097 | 65 | I>V | No | Ensembl | |
| rs774223240 | 67 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs774223240 | 67 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1402393684 | 67 | G>V | No | TOPMed | |
| rs148867971 | 68 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1304482491 | 69 | G>D | No | gnomAD | |
| rs752447530 | 69 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1914792990 | 70 | Y>* | No | TOPMed | |
| rs1293328313 | 70 | Y>H | No | gnomAD | |
|
COSM1392339 rs1914793059 |
71 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| TCGA novel | 71 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779034100 | 72 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1246435230 | 72 | E>K | No |
TOPMed gnomAD |
|
| rs1246435230 | 72 | E>Q | No |
TOPMed gnomAD |
|
| rs779034100 | 72 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs573354292 | 74 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs573354292 | 74 | E>Q | No |
1000Genomes ExAC gnomAD |
|
| rs758911514 | 75 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs1260835156 COSM4687377 |
75 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs747333396 | 76 | K>Q | No |
ExAC gnomAD |
|
| rs1914793833 | 76 | K>R | No | TOPMed | |
| rs1914793905 | 77 | G>D | No |
TOPMed gnomAD |
|
| rs867328918 | 78 | F>L | No | Ensembl | |
| rs1914794130 | 78 | F>S | No | Ensembl | |
| rs1245473019 | 79 | R>G | No |
TOPMed gnomAD |
|
| rs377345709 | 79 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1245473019 | 79 | R>W | No |
TOPMed gnomAD |
|
| rs541029560 | 80 | P>S | No |
1000Genomes ExAC gnomAD |
|
| rs759422477 | 82 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs759422477 | 82 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs759422477 | 82 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs760394355 | 83 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1318539355 | 85 | N>K | No |
TOPMed gnomAD |
|
| rs1309913328 | 86 | I>S | No | TOPMed | |
| rs1408195475 | 86 | I>V | No |
TOPMed gnomAD |
|
| rs140597256 | 88 | V>L | No |
ESP TOPMed gnomAD |
|
| rs140597256 | 88 | V>M | No |
ESP TOPMed gnomAD |
|
| rs1376644489 | 90 | M>I | No | gnomAD | |
| rs1914795414 | 90 | M>T | No | Ensembl | |
| rs757375947 | 91 | R>P | No | TOPMed | |
| rs757375947 | 91 | R>Q | No | TOPMed | |
| rs1307234170 | 92 | A>P | No |
TOPMed gnomAD |
|
| rs1265601037 | 93 | M>I | No | gnomAD | |
| rs373914616 | 93 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1315852699 | 93 | M>V | No | gnomAD | |
| rs1914796053 | 94 | I>L | No | TOPMed | |
| rs1203510941 | 95 | E>G | No | gnomAD | |
| rs1462163503 | 95 | E>K | No | gnomAD | |
| rs1178039894 | 97 | M>I | No | gnomAD | |
| rs564736265 | 97 | M>R | No |
1000Genomes ExAC gnomAD |
|
| rs564736265 | 97 | M>T | No |
1000Genomes ExAC gnomAD |
|
| rs925980060 | 97 | M>V | No |
TOPMed gnomAD |
|
| rs1417467645 | 99 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs765270851 | 99 | R>W | No |
ExAC gnomAD |
|
| rs1470280131 | 102 | I>F | No | gnomAD | |
| rs1914796966 | 102 | I>M | No | TOPMed | |
| rs758854339 | 102 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs1470280131 | 102 | I>V | No | gnomAD | |
| rs145733927 | 103 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1444066811 | 104 | F>S | No |
TOPMed gnomAD |
|
| rs377517036 | 105 | S>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs377517036 | 105 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs755461939 | 105 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1187519069 | 106 | R>W | No | TOPMed | |
| rs748943590 | 108 | E>K | No |
ExAC gnomAD |
|
| rs1261785199 | 109 | S>N | No | gnomAD | |
| rs1253541109 | 110 | K>N | No | TOPMed | |
| rs2144851270 | 110 | K>T | No | Ensembl | |
| rs200110632 | 111 | H>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200110632 | 111 | H>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs141840304 | 111 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1415802898 | 112 | H>Q | No |
TOPMed gnomAD |
|
| rs771240163 | 113 | A>P | No |
ExAC gnomAD |
|
| rs375635151 | 113 | A>V | No |
ESP TOPMed |
|
|
RCV000969111 rs139115108 |
114 | S>N | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs772040139 | 114 | S>R | No |
ExAC gnomAD |
|
| rs774583770 | 114 | S>R | No |
ExAC gnomAD |
|
| rs149877401 | 117 | M>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
TCGA novel rs1914842437 |
117 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 119 | Q>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1914842765 | 121 | P>R | No | TOPMed | |
| rs761059634 | 124 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1400750567 | 126 | T>K | No |
TOPMed gnomAD |
|
| TCGA novel | 126 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764555365 | 128 | E>A | No |
ExAC gnomAD |
|
| rs1159274987 | 129 | K>Q | No | TOPMed | |
| rs371651845 | 130 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs371651845 | 130 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs765960537 | 130 | R>H | No |
ExAC gnomAD |
|
| rs765960537 | 130 | R>L | No |
ExAC gnomAD |
|
| rs751076987 | 131 | Y>* | No |
ExAC gnomAD |
|
| rs902119948 | 131 | Y>C | No |
TOPMed gnomAD |
|
| rs902119948 | 131 | Y>S | No |
TOPMed gnomAD |
|
| rs1242908457 | 132 | A>S | No | gnomAD | |
| rs1351583596 | 133 | A>V | No | gnomAD | |
| rs752729213 | 134 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs752729213 | 134 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1914844186 | 135 | M>I | No | Ensembl | |
| rs1260773989 | 135 | M>L | No | TOPMed | |
| rs62127298 | 136 | Q>K | No | Ensembl | |
| rs1205441758 | 136 | Q>R | No | gnomAD | |
| rs1243554188 | 137 | W>* | No | gnomAD | |
| rs1914844555 | 138 | L>R | No | Ensembl | |
| COSM1564525 | 139 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777656970 | 139 | W>G | No |
ExAC TOPMed gnomAD |
|
| rs749044353 | 140 | R>G | No |
ExAC gnomAD |
|
| rs771281132 | 140 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs1599325501 | 141 | D>G | No | Ensembl | |
| rs779344852 | 141 | D>N | No |
ExAC gnomAD |
|
| rs779344852 | 141 | D>Y | No |
ExAC gnomAD |
|
| rs772239006 | 142 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs772239006 | 142 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1914845429 | 143 | G>S | No | TOPMed | |
| rs369811746 | 145 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3787631 rs1914845604 |
145 | R>Q | pancreas [Cosmic] | No |
cosmic curated TOPMed |
| rs369811746 | 145 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1006352290 | 146 | A>D | No | TOPMed | |
| rs766783580 | 146 | A>T | No | Ensembl | |
| rs310680 | 147 | C>F | No |
1000Genomes ESP ExAC gnomAD |
|
| TCGA novel | 147 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_028000 rs310680 |
147 | C>Y | No |
UniProt 1000Genomes ESP ExAC dbSNP gnomAD |
|
| rs765585779 | 148 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1326276918 | 149 | E>D | No | Ensembl | |
| rs1245753040 | 149 | E>V | No | TOPMed | |
|
rs774052972 COSM1208390 |
150 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs774052972 | 150 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs759088980 | 150 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs752109453 | 151 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1345791697 | 152 | R>L | No | gnomAD | |
| rs1263645841 | 152 | R>W | No |
TOPMed gnomAD |
|
|
rs867791191 COSM3532103 |
153 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs764054033 | 155 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs764054033 | 155 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs980800256 | 157 | L>P | No |
TOPMed gnomAD |
|
|
rs1914847547 COSM1208391 |
158 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs778829713 | 158 | D>V | No |
ExAC gnomAD |
|
| rs746129065 | 159 | S>* | No |
ExAC gnomAD |
|
| rs747001611 | 161 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs747001611 | 161 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1372088870 | 162 | Y>H | No |
TOPMed gnomAD |
|
| rs868282050 | 163 | Y>C | No | Ensembl | |
| rs865832368 | 163 | Y>H | No | Ensembl | |
| rs1914885378 | 166 | H>N | No | TOPMed | |
| COSM994463 | 167 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201656938 | 168 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756566543 | 168 | E>G | No | Ensembl | |
| rs76346810 | 168 | E>K | No | Ensembl | |
| rs199847220 | 169 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs751779470 | 169 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs199847220 | 169 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1187081050 | 170 | I>L | No |
TOPMed gnomAD |
|
| rs755162317 | 171 | T>A | No |
ExAC gnomAD |
|
| rs1445502765 | 171 | T>I | No | gnomAD | |
| rs755162317 | 171 | T>P | No |
ExAC gnomAD |
|
| rs749787028 | 172 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2144855125 | 173 | E>G | No | Ensembl | |
|
COSM108657 rs139079286 |
173 | E>K | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs1914886660 | 174 | G>D | No | Ensembl | |
| rs1478806817 | 174 | G>S | No |
TOPMed gnomAD |
|
|
rs1914886729 COSM3959885 |
175 | Y>C | lung [Cosmic] | No |
cosmic curated TOPMed |
|
rs201355990 COSM4076646 |
176 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1389308286 | 177 | P>L | No |
TOPMed gnomAD |
|
| rs1389308286 | 177 | P>R | No |
TOPMed gnomAD |
|
| rs746672546 | 178 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1318333423 | 179 | A>G | No |
TOPMed gnomAD |
|
| rs1318333423 | 179 | A>V | No |
TOPMed gnomAD |
|
| rs1205782310 | 180 | Q>P | No |
TOPMed gnomAD |
|
| rs1205782310 | 180 | Q>R | No |
TOPMed gnomAD |
|
| rs143953630 | 182 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs761338145 | 184 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs200432733 | 184 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200432733 | 184 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 185 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773083225 | 186 | R>C | No |
ExAC gnomAD |
|
|
rs867130891 COSM710491 |
186 | R>H | lung endometrium central_nervous_system [Cosmic] | No |
cosmic curated Ensembl |
|
rs2230330 RCV000973222 |
187 | M>I | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinVar dbSNP |
|
| rs1599326429 | 187 | M>T | No | gnomAD | |
| rs751333139 | 191 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1914888165 | 191 | G>S | No | gnomAD | |
| rs1914888310 | 192 | I>V | No | Ensembl | |
| rs115644662 | 193 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1914888459 | 193 | N>S | No | Ensembl | |
|
rs775021862 COSM178593 |
194 | E>K | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs1914888964 | 195 | Y>H | No | TOPMed | |
| rs756241789 | 196 | C>R | No |
ExAC gnomAD |
|
| rs1914889207 | 197 | F>S | No | Ensembl | |
| rs1156831129 | 198 | S>P | No |
TOPMed gnomAD |
|
|
rs151214875 COSM1208393 |
199 | V>M | oesophagus large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1914889588 | 200 | Q>E | No | gnomAD | |
| rs369602953 | 201 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs754338447 | 201 | K>Q | No |
ExAC gnomAD |
|
| rs968239084 | 201 | K>R | No | TOPMed | |
| rs1253691718 | 204 | L>V | No | TOPMed | |
| rs1288462652 | 205 | R>Q | No | gnomAD | |
| rs746938649 | 205 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1914999642 | 207 | V>G | No | TOPMed | |
| rs755751411 | 207 | V>M | No |
ExAC gnomAD |
|
| rs367834807 | 208 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 209 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748777205 | 209 | V>I | No |
ExAC gnomAD |
|
| rs770781206 | 210 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs774341405 | 211 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs774341405 | 211 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1377254685 | 213 | K>Q | No | gnomAD | |
| rs771890721 | 216 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs775221783 | 216 | R>H | No |
ExAC gnomAD |
|
| rs1349909924 | 217 | K>* | No | gnomAD | |
| TCGA novel | 217 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3532107 | 219 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs796975912 | 219 | W>C | No | TOPMed | |
| rs1436462432 | 219 | W>G | No | gnomAD | |
| rs1436462432 | 219 | W>R | No | gnomAD | |
| COSM4076647 | 220 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1915001464 | 221 | H>N | No | gnomAD | |
| rs1296430786 | 222 | C>R | No |
TOPMed gnomAD |
|
| rs1915001855 | 224 | E>D | No | Ensembl | |
| rs1915002017 | 226 | V>E | No | Ensembl | |
| rs1915002193 | 227 | I>N | No | Ensembl | |
| rs1365825471 | 230 | I>T | No |
TOPMed gnomAD |
|
| rs764319987 | 230 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2144859641 | 231 | Y>C | No | Ensembl | |
| rs1295511630 | 232 | L>P | No |
TOPMed gnomAD |
|
| rs1295511630 | 232 | L>R | No |
TOPMed gnomAD |
|
| rs762017781 | 236 | S>G | No |
ExAC gnomAD |
|
| rs1915003388 | 236 | S>N | No | Ensembl | |
| rs146483408 | 238 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 239 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM5389308 | 239 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375791169 | 241 | C>Y | No |
ESP TOPMed gnomAD |
|
| rs1220789775 | 242 | L>P | No | gnomAD | |
| rs1374119223 | 242 | L>V | No |
TOPMed gnomAD |
|
| TCGA novel | 243 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1029823852 | 244 | E>* | No |
TOPMed gnomAD |
|
| TCGA novel | 244 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1915003870 | 244 | E>G | No | Ensembl | |
| rs1029823852 | 244 | E>K | No |
TOPMed gnomAD |
|
| rs1029823852 | 244 | E>Q | No |
TOPMed gnomAD |
|
| rs1443850109 | 246 | N>H | No |
TOPMed gnomAD |
|
| rs1915004065 | 246 | N>S | No | gnomAD | |
| rs1011689952 | 247 | Q>R | No |
TOPMed gnomAD |
|
| TCGA novel | 248 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3718248 rs200649442 |
250 | R>C | liver [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs773385959 | 250 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs145665648 | 251 | M>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1252636922 | 251 | M>R | No |
TOPMed gnomAD |
|
| rs145665648 | 251 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1351158900 | 252 | K>Q | No | gnomAD | |
|
COSM4154156 rs199709383 |
254 | S>R | kidney [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs760016259 | 256 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 258 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1395054270 | 259 | G>V | No |
TOPMed gnomAD |
|
| rs1656782963 | 260 | T>S | No | TOPMed | |
| TCGA novel | 261 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1915062291 | 262 | L>Q | No | TOPMed | |
| TCGA novel | 263 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4831239 | 263 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1441521059 | 263 | E>K | No | gnomAD | |
| rs767761894 | 265 | P>T | No |
ExAC gnomAD |
|
| rs1915062623 | 266 | W>S | No | TOPMed | |
| rs753564320 | 268 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs756817009 | 269 | S>N | No |
ExAC gnomAD |
|
| COSM418342 | 270 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1304349285 | 271 | S>F | No |
TOPMed gnomAD |
|
| rs1481671679 | 272 | V>I | No | gnomAD | |
| rs1173531250 | 273 | I>V | No | gnomAD | |
|
COSM4404302 rs1915063259 |
274 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs749873249 | 274 | L>R | No |
ExAC gnomAD |
|
| rs1474419176 | 278 | K>N | No | gnomAD | |
| rs1915063693 | 279 | T>S | No | Ensembl | |
| rs1156779924 | 280 | D>N | No |
TOPMed gnomAD |
|
| rs746905865 | 282 | L>P | No |
ExAC gnomAD |
|
| rs143247880 | 283 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1915064470 | 284 | E>* | No | Ensembl | |
| rs1915064622 | 286 | I>V | No |
TOPMed gnomAD |
|
| rs1275247896 | 287 | P>L | No |
TOPMed gnomAD |
|
|
rs868247802 COSM3532108 |
287 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1198409269 | 288 | T>A | No | gnomAD | |
| rs773440731 | 288 | T>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 288 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1483130427 | 289 | S>F | No |
TOPMed gnomAD |
|
| rs1483130427 | 289 | S>Y | No |
TOPMed gnomAD |
|
| rs1266791223 | 290 | H>Q | No |
TOPMed gnomAD |
|
| rs1203827962 | 292 | A>S | No | gnomAD | |
| rs1203827962 | 292 | A>T | No | gnomAD | |
| rs1248897452 | 293 | T>A | No | gnomAD | |
| rs1200174902 | 294 | Y>C | No | gnomAD | |
| rs763062805 | 295 | F>I | No |
ExAC gnomAD |
|
| rs1330649492 | 296 | P>L | No |
TOPMed gnomAD |
|
| rs759941706 | 299 | Q>* | No |
ExAC gnomAD |
|
| rs1915066345 | 299 | Q>R | No | TOPMed | |
| rs1555707385 | 300 | G>R | No | gnomAD | |
| rs1555707385 | 300 | G>S | No | gnomAD | |
| rs1446504412 | 300 | G>V | No | gnomAD | |
| rs1272031360 | 301 | P>L | No |
TOPMed gnomAD |
|
| COSM142295 | 301 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1357304874 | 302 | K>E | No |
TOPMed gnomAD |
|
| rs774252847 | 302 | K>N | No |
ExAC gnomAD |
|
| rs1915169237 | 302 | K>R | No | TOPMed | |
| rs1915169386 | 304 | D>Y | No | Ensembl | |
| rs1017132181 | 308 | A>T | No | Ensembl | |
| COSM4849707 | 309 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746406189 | 309 | K>Q | No |
ExAC gnomAD |
|
| rs772397465 | 309 | K>R | No |
ExAC gnomAD |
|
| rs1568300677 | 310 | R>K | No | Ensembl | |
| rs963088906 | 311 | F>L | No | Ensembl | |
| rs2144866821 | 312 | I>T | No | Ensembl | |
| rs1324531419 | 313 | L>P | No |
TOPMed gnomAD |
|
| rs775924021 | 313 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs761093347 | 314 | D>V | No |
ExAC gnomAD |
|
| rs1354906492 | 315 | M>K | No |
TOPMed gnomAD |
|
| rs1915170059 | 315 | M>L | No |
TOPMed gnomAD |
|
| rs201928137 | 317 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1393781581 | 317 | T>S | No | TOPMed | |
| rs1599332869 | 320 | Y>H | No | Ensembl | |
| rs955365992 | 321 | T>I | No |
TOPMed gnomAD |
|
| rs955365992 | 321 | T>N | No |
TOPMed gnomAD |
|
| rs762561049 | 321 | T>P | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs372928786 |
322 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC TOPMed gnomAD NCI-TCGA |
| rs1915171324 | 323 | C>Y | No |
TOPMed gnomAD |
|
| rs754957377 | 324 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs767291635 | 325 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs752679814 | 326 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1389192934 | 326 | G>D | No | gnomAD | |
| rs752679814 | 326 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs752679814 | 326 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs777690665 | 327 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs757502333 | 328 | E>D | No |
ExAC TOPMed gnomAD |
|
|
rs1396214838 COSM123390 |
328 | E>K | upper_aerodigestive_tract Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs745864047 | 332 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs772184102 | 332 | K>R | No |
ExAC gnomAD |
|
| rs747491972 | 334 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs747491972 | 334 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs747491972 | 334 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1483718966 | 334 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1465632602 | 335 | R>* | No | Ensembl | |
| rs1915172756 | 335 | R>L | No | Ensembl | |
| rs867088858 | 338 | R>C | No |
TOPMed gnomAD |
|
| rs1184334208 | 338 | R>H | No | gnomAD | |
| rs867088858 | 338 | R>S | No |
TOPMed gnomAD |
|
| rs1028072297 | 339 | L>F | No | TOPMed | |
| rs1599332950 | 340 | F>S | No | Ensembl | |
| rs1337172056 | 342 | H>R | No |
TOPMed gnomAD |
|
| rs765889898 | 342 | H>Y | No |
ExAC gnomAD |
|
| rs1360637442 | 343 | Y>C | No |
TOPMed gnomAD |
|
| rs2144866959 | 344 | T>I | No | Ensembl | |
| rs1271750566 | 344 | T>S | No |
TOPMed gnomAD |
|
| rs544907753 | 346 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs752734019 | 349 | T>A | No |
ExAC gnomAD |
|
| rs764576537 | 350 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs764000592 | 353 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs764000592 | 353 | R>G | No |
ExAC TOPMed gnomAD |
|
| COSM3692602 | 353 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764000592 | 353 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2144866997 | 356 | F>L | No | Ensembl | |
| rs1915174393 | 356 | F>V | No | gnomAD | |
| rs867394387 | 357 | K>N | No | Ensembl | |
| rs1232858885 | 357 | K>R | No |
TOPMed gnomAD |
|
| rs757501367 | 359 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs2144867011 | 360 | R>Q | No | Ensembl | |
| rs1205964029 | 360 | R>W | No |
TOPMed gnomAD |
|
| rs1816137028 | 362 | S>L | No | TOPMed | |
| rs1482702897 | 363 | V>L | No |
TOPMed gnomAD |
|
| rs1482702897 | 363 | V>M | No |
TOPMed gnomAD |
|
| rs779067884 | 364 | L>F | No |
ExAC gnomAD |
|
| rs779067884 | 364 | L>V | No |
ExAC gnomAD |
|
| rs1433555721 | 365 | A>S | No |
TOPMed gnomAD |
|
| rs1433555721 | 365 | A>T | No |
TOPMed gnomAD |
|
|
COSM107599 rs144051973 |
366 | R>C | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs144051973 | 366 | R>S | No |
TOPMed gnomAD |
|
| rs758456407 | 369 | D>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 369 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1394097918 | 371 | I>V | No | TOPMed | |
| rs780149273 | 372 | N>H | No |
ExAC gnomAD |
|
| rs575093003 | 372 | N>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs575093003 | 372 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1440710887 | 373 | L>V | No | gnomAD |
No associated diseases with P30679
No regional properties for P30679
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P30679 | |||
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| action potential | A process in which membrane potential cycles through a depolarizing spike, triggered in response to depolarization above some threshold, followed by repolarization. This cycle is driven by the flow of ions through various voltage gated channels with different thresholds and ion specificities. |
| activation of phospholipase C activity | The initiation of the activity of the inactive enzyme phospolipase C as the result of The series of molecular signals generated as a consequence of a G protein-coupled receptor binding to its physiological ligand. |
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| calcium-mediated signaling | Any intracellular signal transduction in which the signal is passed on within the cell via calcium ions. |
| phospholipase C-activating dopamine receptor signaling pathway | A phospholipase C-activating receptor G protein-coupled receptor signaling pathway initiated by dopamine binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | A phospholipase C-activating G protein-coupled receptor signaling pathway initiated by acetylcholine binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38408 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Bos taurus (Bovine) | PR |
| P23625 | Galphaq | G protein alpha q subunit | Drosophila melanogaster (Fruit fly) | PR |
| O95837 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Homo sapiens (Human) | SS |
| P29992 | GNA11 | Guanine nucleotide-binding protein subunit alpha-11 | Homo sapiens (Human) | SS |
| P50148 | GNAQ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P11488 | GNAT1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| P38405 | GNAL | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P63096 | GNAI1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | EV |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P21279 | Gnaq | Guanine nucleotide-binding protein G(q) subunit alpha | Mus musculus (Mouse) | PR |
| P30678 | Gna15 | Guanine nucleotide-binding protein subunit alpha-15 | Mus musculus (Mouse) | PR |
| Q2PKF4 | GNAQ | Guanine nucleotide-binding protein G(q) subunit alpha | Sus scrofa (Pig) | PR |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| Q9JID2 | Gna11 | Guanine nucleotide-binding protein subunit alpha-11 | Rattus norvegicus (Rat) | SS |
| P82471 | Gnaq | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| O88302 | Gna15 | Guanine nucleotide-binding protein subunit alpha-15 | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARSLTWRCC | PWCLTEDEKA | AARVDQEINR | ILLEQKKQDR | GELKLLLLGP | GESGKSTFIK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QMRIIHGAGY | SEEERKGFRP | LVYQNIFVSM | RAMIEAMERL | QIPFSRPESK | HHASLVMSQD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PYKVTTFEKR | YAAAMQWLWR | DAGIRACYER | RREFHLLDSA | VYYLSHLERI | TEEGYVPTAQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DVLRSRMPTT | GINEYCFSVQ | KTNLRIVDVG | GQKSERKKWI | HCFENVIALI | YLASLSEYDQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CLEENNQENR | MKESLALFGT | ILELPWFKST | SVILFLNKTD | ILEEKIPTSH | LATYFPSFQG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PKQDAEAAKR | FILDMYTRMY | TGCVDGPEGS | KKGARSRRLF | SHYTCATDTQ | NIRKVFKDVR |
| 370 | |||||
| DSVLARYLDE | INLL |