P29476
Gene name |
Nos1 (Bnos) |
Protein name |
Nitric oxide synthase, brain |
Names |
BNOS, Constitutive NOS, NC-NOS, NOS type I, Neuronal NOS, N-NOS, nNOS, Peptidyl-cysteine S-nitrosylase NOS1 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:24598 |
EC number |
1.14.13.39: With NADH or NADPH as one donor, and incorporation of one atom of oxygen |
Protein Class |
|
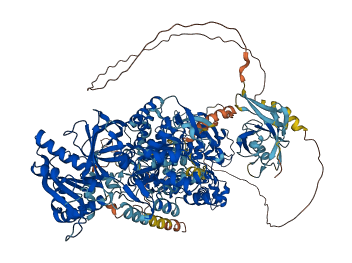
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
351-712 (NO synthase domain); 990-1358 (FAD/NAD-binding domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

318 structures for P29476
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1B8Q | NMR | - | A | 11-133 | PDB |
| 1CMI | X-ray | 250 A | C/D | 225-237 | PDB |
| 1F20 | X-ray | 190 A | A | 963-1397 | PDB |
| 1K2R | X-ray | 215 A | A/B | 299-717 | PDB |
| 1K2S | X-ray | 255 A | A/B | 299-717 | PDB |
| 1K2T | X-ray | 220 A | A/B | 299-717 | PDB |
| 1K2U | X-ray | 220 A | A/B | 299-717 | PDB |
| 1LZX | X-ray | 200 A | A/B | 299-717 | PDB |
| 1LZZ | X-ray | 205 A | A/B | 299-717 | PDB |
| 1M00 | X-ray | 205 A | A/B | 299-717 | PDB |
| 1MMV | X-ray | 200 A | A/B | 299-717 | PDB |
| 1MMW | X-ray | 200 A | A/B | 299-717 | PDB |
| 1OM4 | X-ray | 175 A | A/B | 297-718 | PDB |
| 1OM5 | X-ray | 230 A | A/B | 297-717 | PDB |
| 1P6H | X-ray | 198 A | A/B | 297-717 | PDB |
| 1P6I | X-ray | 190 A | A/B | 297-717 | PDB |
| 1P6J | X-ray | 200 A | A/B | 297-717 | PDB |
| 1P6K | X-ray | 178 A | A/B | 297-717 | PDB |
| 1QAU | X-ray | 125 A | A | 14-125 | PDB |
| 1QAV | X-ray | 190 A | B | 12-126 | PDB |
| 1QW6 | X-ray | 210 A | A | 298-716 | PDB |
| 1QWC | X-ray | 230 A | A | 298-716 | PDB |
| 1RS6 | X-ray | 195 A | A/B | 297-717 | PDB |
| 1RS7 | X-ray | 195 A | A/B | 297-717 | PDB |
| 1TLL | X-ray | 230 A | A/B | 742-1429 | PDB |
| 1VAG | X-ray | 200 A | A | 298-716 | PDB |
| 1ZVI | X-ray | 200 A | A | 298-716 | PDB |
| 1ZVL | X-ray | 250 A | A/B | 298-716 | PDB |
| 1ZZQ | X-ray | 190 A | A/B | 299-718 | PDB |
| 1ZZR | X-ray | 205 A | A/B | 299-718 | PDB |
| 1ZZU | X-ray | 190 A | A/B | 299-718 | PDB |
| 2G6H | X-ray | 200 A | A/B | 299-718 | PDB |
| 2G6I | X-ray | 190 A | A/B | 299-718 | PDB |
| 2G6J | X-ray | 230 A | A/B | 299-718 | PDB |
| 2G6K | X-ray | 200 A | A/B | 299-718 | PDB |
| 2G6L | X-ray | 205 A | A/B | 299-718 | PDB |
| 2G6M | X-ray | 185 A | A/B | 299-718 | PDB |
| 2G6N | X-ray | 190 A | A/B | 299-718 | PDB |
| 2HX3 | X-ray | 200 A | A/B | 297-718 | PDB |
| 2HX4 | X-ray | 215 A | A/B | 297-718 | PDB |
| 3B3M | X-ray | 195 A | A/B | 297-718 | PDB |
| 3B3N | X-ray | 198 A | A/B | 297-718 | PDB |
| 3B3O | X-ray | 205 A | A/B | 297-718 | PDB |
| 3B3P | X-ray | 245 A | A/B | 297-718 | PDB |
| 3DQR | X-ray | 240 A | A/B | 297-718 | PDB |
| 3FC5 | X-ray | 259 A | A/B | 297-718 | PDB |
| 3HSN | X-ray | 191 A | A/B | 297-718 | PDB |
| 3HSO | X-ray | 202 A | A/B | 297-718 | PDB |
| 3HSP | X-ray | 220 A | A/B | 297-718 | PDB |
| 3JT3 | X-ray | 215 A | A/B | 297-718 | PDB |
| 3JT4 | X-ray | 180 A | A/B | 297-718 | PDB |
| 3JT5 | X-ray | 210 A | A/B | 297-718 | PDB |
| 3JT6 | X-ray | 220 A | A/B | 297-718 | PDB |
| 3JT7 | X-ray | 210 A | A/B | 297-718 | PDB |
| 3JT8 | X-ray | 195 A | A/B | 297-718 | PDB |
| 3JT9 | X-ray | 210 A | A/B | 297-718 | PDB |
| 3JTA | X-ray | 218 A | A/B | 297-718 | PDB |
| 3JWS | X-ray | 195 A | A/B | 297-718 | PDB |
| 3JWT | X-ray | 201 A | A/B | 297-718 | PDB |
| 3JWU | X-ray | 193 A | A/B | 297-718 | PDB |
| 3JWV | X-ray | 198 A | A/B | 297-718 | PDB |
| 3JX0 | X-ray | 220 A | A/B | 297-718 | PDB |
| 3JX1 | X-ray | 200 A | A/B | 297-718 | PDB |
| 3JX2 | X-ray | 210 A | A/B | 297-718 | PDB |
| 3JX3 | X-ray | 195 A | A/B | 297-718 | PDB |
| 3JX4 | X-ray | 226 A | A/B | 297-718 | PDB |
| 3JX5 | X-ray | 215 A | A/B | 297-718 | PDB |
| 3JX6 | X-ray | 235 A | A/B | 297-718 | PDB |
| 3N2R | X-ray | 190 A | A/B | 297-718 | PDB |
| 3N5V | X-ray | 230 A | A/B | 297-718 | PDB |
| 3N5W | X-ray | 173 A | A/B | 297-718 | PDB |
| 3N5X | X-ray | 180 A | A/B | 297-718 | PDB |
| 3N5Y | X-ray | 205 A | A/B | 297-718 | PDB |
| 3N5Z | X-ray | 218 A | A/B | 297-718 | PDB |
| 3N60 | X-ray | 198 A | A/B | 297-718 | PDB |
| 3N61 | X-ray | 195 A | A/B | 297-718 | PDB |
| 3N62 | X-ray | 195 A | A/B | 297-718 | PDB |
| 3N63 | X-ray | 200 A | A/B | 297-718 | PDB |
| 3N64 | X-ray | 195 A | A/B | 297-718 | PDB |
| 3N65 | X-ray | 180 A | A/B | 297-718 | PDB |
| 3N66 | X-ray | 178 A | A/B | 297-718 | PDB |
| 3N67 | X-ray | 209 A | A/B | 298-711 | PDB |
| 3N68 | X-ray | 253 A | A/B | 298-711 | PDB |
| 3N69 | X-ray | 265 A | A/B | 298-711 | PDB |
| 3N6A | X-ray | 249 A | A/B | 298-711 | PDB |
| 3N6B | X-ray | 310 A | A/B | 298-711 | PDB |
| 3N6C | X-ray | 306 A | A/B | 298-711 | PDB |
| 3N6D | X-ray | 305 A | A/B | 298-711 | PDB |
| 3N6E | X-ray | 220 A | A/B | 298-711 | PDB |
| 3N6F | X-ray | 218 A | A/B | 298-711 | PDB |
| 3N6G | X-ray | 221 A | A/B | 298-711 | PDB |
| 3NLJ | X-ray | 220 A | A/B | 297-718 | PDB |
| 3NLK | X-ray | 202 A | A/B | 297-718 | PDB |
| 3NLM | X-ray | 185 A | A/B | 297-718 | PDB |
| 3NLN | X-ray | 200 A | A/B | 297-718 | PDB |
| 3NLO | X-ray | 230 A | A/B | 297-718 | PDB |
| 3NLP | X-ray | 202 A | A/B | 297-718 | PDB |
| 3NLQ | X-ray | 215 A | A/B | 297-718 | PDB |
| 3NLR | X-ray | 210 A | A/B | 297-718 | PDB |
| 3NLV | X-ray | 210 A | A/B | 297-718 | PDB |
| 3NLW | X-ray | 210 A | A/B | 297-718 | PDB |
| 3NLX | X-ray | 187 A | A/B | 297-718 | PDB |
| 3NLY | X-ray | 199 A | A/B | 297-718 | PDB |
| 3NLZ | X-ray | 192 A | A/B | 297-718 | PDB |
| 3NM0 | X-ray | 181 A | A/B | 297-718 | PDB |
| 3NNY | X-ray | 210 A | A/B | 297-718 | PDB |
| 3NNZ | X-ray | 197 A | A/B | 297-718 | PDB |
| 3PNE | X-ray | 197 A | A/B | 297-718 | PDB |
| 3PNF | X-ray | 194 A | A/B | 297-718 | PDB |
| 3PNG | X-ray | 188 A | A/B | 297-718 | PDB |
| 3Q99 | X-ray | 215 A | A/B | 297-718 | PDB |
| 3Q9A | X-ray | 224 A | A/B | 297-718 | PDB |
| 3RQJ | X-ray | 184 A | A/B | 297-718 | PDB |
| 3RQK | X-ray | 221 A | A/B | 297-718 | PDB |
| 3RQL | X-ray | 193 A | A/B | 297-718 | PDB |
| 3RQM | X-ray | 195 A | A/B | 297-718 | PDB |
| 3RQN | X-ray | 195 A | A/B | 297-718 | PDB |
| 3SVP | X-ray | 205 A | A/B | 297-718 | PDB |
| 3SVQ | X-ray | 218 A | A/B | 297-718 | PDB |
| 3TYL | X-ray | 190 A | A/B | 297-718 | PDB |
| 3TYM | X-ray | 200 A | A/B | 297-718 | PDB |
| 3TYN | X-ray | 197 A | A/B | 297-718 | PDB |
| 3TYO | X-ray | 193 A | A/B | 297-718 | PDB |
| 3UFO | X-ray | 217 A | A/B | 297-718 | PDB |
| 3UFP | X-ray | 210 A | A/B | 297-718 | PDB |
| 3UFQ | X-ray | 206 A | A/B | 297-718 | PDB |
| 3UFR | X-ray | 210 A | A/B | 297-718 | PDB |
| 3UFS | X-ray | 197 A | A/B | 297-718 | PDB |
| 3UFT | X-ray | 208 A | A/B | 297-718 | PDB |
| 3UFU | X-ray | 189 A | A/B | 297-718 | PDB |
| 3UFV | X-ray | 208 A | A/B | 297-718 | PDB |
| 3UFW | X-ray | 200 A | A/B | 297-718 | PDB |
| 4C39 | X-ray | 198 A | A/B | 297-718 | PDB |
| 4CAM | X-ray | 183 A | A/B | 297-718 | PDB |
| 4CAN | X-ray | 191 A | A/B | 297-718 | PDB |
| 4CAO | X-ray | 198 A | A/B | 297-718 | PDB |
| 4CAP | X-ray | 206 A | A/B | 297-718 | PDB |
| 4CAQ | X-ray | 195 A | A/B | 297-718 | PDB |
| 4CDT | X-ray | 200 A | A/B | 297-718 | PDB |
| 4CTP | X-ray | 205 A | A/B | 297-718 | PDB |
| 4CTQ | X-ray | 200 A | A/B | 297-718 | PDB |
| 4CTR | X-ray | 220 A | A/B | 297-718 | PDB |
| 4CTT | X-ray | 230 A | A/B | 297-718 | PDB |
| 4CTU | X-ray | 216 A | A/B | 297-718 | PDB |
| 4CTV | X-ray | 178 A | A/B | 297-718 | PDB |
| 4CTW | X-ray | 190 A | A/B | 297-718 | PDB |
| 4CTX | X-ray | 182 A | A/B | 297-718 | PDB |
| 4CX3 | X-ray | 197 A | A/B | 297-718 | PDB |
| 4CX4 | X-ray | 198 A | A/B | 297-718 | PDB |
| 4CX5 | X-ray | 180 A | A/B | 297-718 | PDB |
| 4CX6 | X-ray | 190 A | A/B | 297-718 | PDB |
| 4D2Y | X-ray | 198 A | A/B | 297-718 | PDB |
| 4D2Z | X-ray | 189 A | A/B | 297-718 | PDB |
| 4D30 | X-ray | 196 A | A/B | 297-718 | PDB |
| 4D31 | X-ray | 195 A | A/B | 297-718 | PDB |
| 4D32 | X-ray | 210 A | A/B | 297-718 | PDB |
| 4D3B | X-ray | 180 A | A/B | 297-718 | PDB |
| 4D7O | X-ray | 178 A | A/B | 297-718 | PDB |
| 4EUX | X-ray | 214 A | A/B | 297-718 | PDB |
| 4FVW | X-ray | 181 A | A/B | 297-718 | PDB |
| 4FVX | X-ray | 200 A | A/B | 297-718 | PDB |
| 4FVY | X-ray | 170 A | A/B | 297-718 | PDB |
| 4FVZ | X-ray | 199 A | A/B | 297-718 | PDB |
| 4FW0 | X-ray | 195 A | A/B | 297-718 | PDB |
| 4GQE | X-ray | 180 A | A/B | 297-718 | PDB |
| 4HOP | X-ray | 229 A | B/D/F | 4-126 | PDB |
| 4IMS | X-ray | 215 A | A/B | 297-718 | PDB |
| 4IMT | X-ray | 220 A | A/B | 297-718 | PDB |
| 4IMU | X-ray | 203 A | A/B | 297-718 | PDB |
| 4IMW | X-ray | 220 A | A/B | 297-718 | PDB |
| 4JSE | X-ray | 197 A | A/B | 297-718 | PDB |
| 4JSF | X-ray | 205 A | A/B | 297-718 | PDB |
| 4JSG | X-ray | 194 A | A/B | 297-718 | PDB |
| 4JSH | X-ray | 235 A | A/B | 297-718 | PDB |
| 4JSI | X-ray | 209 A | A/B | 297-718 | PDB |
| 4JSJ | X-ray | 192 A | A/B | 297-718 | PDB |
| 4K5D | X-ray | 210 A | A/B | 297-718 | PDB |
| 4K5E | X-ray | 189 A | A/B | 297-718 | PDB |
| 4K5F | X-ray | 220 A | A/B | 297-718 | PDB |
| 4K5G | X-ray | 185 A | A/B | 297-718 | PDB |
| 4KCH | X-ray | 215 A | A/B | 297-718 | PDB |
| 4KCI | X-ray | 227 A | A/B | 297-718 | PDB |
| 4KCJ | X-ray | 205 A | A/B | 297-718 | PDB |
| 4KCK | X-ray | 210 A | A/B | 297-718 | PDB |
| 4KCL | X-ray | 193 A | A/B | 297-718 | PDB |
| 4KCM | X-ray | 207 A | A/B | 297-718 | PDB |
| 4KCN | X-ray | 185 A | A/B | 297-718 | PDB |
| 4KCO | X-ray | 186 A | A/B | 297-718 | PDB |
| 4LUX | X-ray | 186 A | A/B | 297-718 | PDB |
| 4UGZ | X-ray | 208 A | A/B | 297-718 | PDB |
| 4UH0 | X-ray | 204 A | A/B | 297-718 | PDB |
| 4UH1 | X-ray | 180 A | A/B | 297-718 | PDB |
| 4UH2 | X-ray | 199 A | A/B | 297-718 | PDB |
| 4UH3 | X-ray | 203 A | A/B | 297-718 | PDB |
| 4UH4 | X-ray | 195 A | A/B | 297-718 | PDB |
| 4UPM | X-ray | 190 A | A/B | 297-718 | PDB |
| 4UPN | X-ray | 209 A | A/B | 297-718 | PDB |
| 4UPO | X-ray | 195 A | A/B | 297-718 | PDB |
| 4UPP | X-ray | 191 A | A/B | 297-718 | PDB |
| 4V3V | X-ray | 206 A | A/B | 297-718 | PDB |
| 4V3W | X-ray | 213 A | A/B | 297-718 | PDB |
| 4V3X | X-ray | 199 A | A/B | 297-718 | PDB |
| 4V3Y | X-ray | 196 A | A/B | 297-718 | PDB |
| 4V3Z | X-ray | 205 A | A/B | 297-718 | PDB |
| 5AD4 | X-ray | 198 A | A/B | 297-718 | PDB |
| 5AD5 | X-ray | 190 A | A/B | 297-718 | PDB |
| 5AD6 | X-ray | 200 A | A/B | 297-718 | PDB |
| 5AD7 | X-ray | 195 A | A/B | 297-718 | PDB |
| 5AD8 | X-ray | 191 A | A/B | 297-718 | PDB |
| 5AD9 | X-ray | 230 A | A/B | 297-718 | PDB |
| 5ADA | X-ray | 198 A | A/B | 297-718 | PDB |
| 5ADB | X-ray | 205 A | A/B | 297-718 | PDB |
| 5ADC | X-ray | 210 A | A/B | 297-718 | PDB |
| 5ADD | X-ray | 210 A | A/B | 297-718 | PDB |
| 5ADE | X-ray | 210 A | A/B | 297-718 | PDB |
| 5AGK | X-ray | 200 A | A/B | 297-718 | PDB |
| 5AGL | X-ray | 194 A | A/B | 297-718 | PDB |
| 5AGM | X-ray | 184 A | A/B | 297-718 | PDB |
| 5AGN | X-ray | 195 A | A/B | 297-718 | PDB |
| 5AGO | X-ray | 190 A | A/B | 297-718 | PDB |
| 5AGP | X-ray | 210 A | A/B | 297-718 | PDB |
| 5FVO | X-ray | 212 A | A | 297-718 | PDB |
| 5FVP | X-ray | 210 A | A/B | 297-718 | PDB |
| 5FVQ | X-ray | 195 A | A/B | 297-718 | PDB |
| 5FVR | X-ray | 184 A | A/B | 297-718 | PDB |
| 5FVS | X-ray | 195 A | A/B | 297-718 | PDB |
| 5FVT | X-ray | 183 A | A/B | 297-718 | PDB |
| 5FW0 | X-ray | 180 A | A/B | 297-718 | PDB |
| 5G0N | X-ray | 194 A | A/B | 297-718 | PDB |
| 5G0O | X-ray | 185 A | A/B | 297-718 | PDB |
| 5G0P | X-ray | 210 A | A/B | 297-718 | PDB |
| 5UNR | X-ray | 195 A | A/B | 297-718 | PDB |
| 5UNS | X-ray | 190 A | A/B | 297-718 | PDB |
| 5UNT | X-ray | 205 A | A/B | 297-718 | PDB |
| 5UNU | X-ray | 205 A | A/B | 297-718 | PDB |
| 5UNV | X-ray | 200 A | A/B | 297-718 | PDB |
| 5UNW | X-ray | 204 A | A/B | 297-718 | PDB |
| 5UNX | X-ray | 203 A | A/B | 297-718 | PDB |
| 5UNY | X-ray | 182 A | A/B | 297-718 | PDB |
| 5UNZ | X-ray | 195 A | A/B | 297-718 | PDB |
| 5UO0 | X-ray | 197 A | A/B | 297-718 | PDB |
| 5VUI | X-ray | 206 A | A/B | 297-718 | PDB |
| 5VUJ | X-ray | 195 A | A/B | 297-718 | PDB |
| 5VUK | X-ray | 195 A | A/B | 297-718 | PDB |
| 5VUL | X-ray | 200 A | A/B | 297-718 | PDB |
| 5VUM | X-ray | 210 A | A/B | 297-718 | PDB |
| 5VUN | X-ray | 175 A | A/B | 297-718 | PDB |
| 5VUO | X-ray | 180 A | A/B | 297-718 | PDB |
| 5VUP | X-ray | 194 A | A/B | 297-718 | PDB |
| 5VUQ | X-ray | 200 A | A/B | 297-718 | PDB |
| 5VUR | X-ray | 197 A | A/B | 297-718 | PDB |
| 5VUS | X-ray | 195 A | A/B | 297-718 | PDB |
| 5VUT | X-ray | 200 A | A/B | 297-718 | PDB |
| 5VUU | X-ray | 196 A | A/B | 297-718 | PDB |
| 6AUQ | X-ray | 195 A | A/B | 297-718 | PDB |
| 6AUR | X-ray | 175 A | A/B | 297-718 | PDB |
| 6AUS | X-ray | 170 A | A/B | 297-718 | PDB |
| 6AUT | X-ray | 190 A | A/B | 297-718 | PDB |
| 6AUU | X-ray | 185 A | A/B | 297-718 | PDB |
| 6AUV | X-ray | 176 A | A/B | 297-718 | PDB |
| 6AUW | X-ray | 170 A | A/B | 297-718 | PDB |
| 6AUX | X-ray | 190 A | A/B | 297-718 | PDB |
| 6NGJ | X-ray | 176 A | A/B | 297-718 | PDB |
| 6NGK | X-ray | 183 A | A/B | 297-718 | PDB |
| 6NGL | X-ray | 183 A | A/B | 297-718 | PDB |
| 6NGM | X-ray | 169 A | A/B | 297-718 | PDB |
| 6NGN | X-ray | 190 A | A/B | 297-718 | PDB |
| 6NGP | X-ray | 198 A | A/B | 297-718 | PDB |
| 6NGQ | X-ray | 195 A | A/B | 297-718 | PDB |
| 6NGR | X-ray | 182 A | A/B | 297-718 | PDB |
| 6NGS | X-ray | 190 A | A/B | 297-718 | PDB |
| 6NGT | X-ray | 194 A | A/B | 297-718 | PDB |
| 6NGU | X-ray | 200 A | A/B | 297-718 | PDB |
| 6NGV | X-ray | 183 A | A/B | 297-718 | PDB |
| 6NGW | X-ray | 186 A | A/B | 297-718 | PDB |
| 6NGX | X-ray | 177 A | A/B | 297-718 | PDB |
| 6NGY | X-ray | 193 A | A/B | 297-718 | PDB |
| 6NGZ | X-ray | 200 A | A/B | 297-718 | PDB |
| 6NH0 | X-ray | 190 A | A/B | 297-718 | PDB |
| 6NHD | X-ray | 210 A | A/B | 297-718 | PDB |
| 6NHE | X-ray | 200 A | A/B | 297-718 | PDB |
| 6PMV | X-ray | 180 A | A/B | 297-718 | PDB |
| 6PMW | X-ray | 175 A | A/B | 297-718 | PDB |
| 6PMX | X-ray | 205 A | A/B | 297-718 | PDB |
| 6PMY | X-ray | 195 A | A/B | 297-718 | PDB |
| 6PMZ | X-ray | 210 A | A/B | 297-718 | PDB |
| 6PN0 | X-ray | 223 A | A/B | 297-718 | PDB |
| 6PN1 | X-ray | 220 A | A/B | 297-718 | PDB |
| 6PN2 | X-ray | 188 A | A/B | 297-718 | PDB |
| 6PN3 | X-ray | 180 A | A/B | 297-718 | PDB |
| 6PN4 | X-ray | 190 A | A/B | 297-718 | PDB |
| 6PN5 | X-ray | 170 A | A/B | 297-718 | PDB |
| 6PN6 | X-ray | 184 A | A/B | 297-718 | PDB |
| 6PN7 | X-ray | 188 A | A/B | 297-718 | PDB |
| 6PN8 | X-ray | 184 A | A/B | 297-718 | PDB |
| 6PN9 | X-ray | 184 A | A/B | 297-718 | PDB |
| 7S3X | X-ray | 195 A | A/B | 297-718 | PDB |
| 7S3Y | X-ray | 208 A | A/B | 297-718 | PDB |
| 7S3Z | X-ray | 173 A | A/B | 297-718 | PDB |
| 7S40 | X-ray | 180 A | A/B | 297-718 | PDB |
| 7TS9 | X-ray | 185 A | A | 297-717 | PDB |
| 7TSA | X-ray | 203 A | A | 297-718 | PDB |
| 7TSB | X-ray | 182 A | A | 297-717 | PDB |
| 7TSC | X-ray | 180 A | A | 297-718 | PDB |
| 7TSD | X-ray | 177 A | A | 297-718 | PDB |
| 7TSE | X-ray | 185 A | A | 297-718 | PDB |
| 7TSF | X-ray | 178 A | A/B | 297-718 | PDB |
| 7UAN | X-ray | 170 A | A | 297-718 | PDB |
| 8FG9 | X-ray | 195 A | A/B | 297-718 | PDB |
| 8FGA | X-ray | 189 A | A/B | 297-718 | PDB |
| 8FGB | X-ray | 191 A | A/B | 297-718 | PDB |
| 8FGC | X-ray | 178 A | A/B | 297-718 | PDB |
| 8FGD | X-ray | 178 A | A | 297-718 | PDB |
| 8FGE | X-ray | 189 A | A | 297-718 | PDB |
| 8FGV | X-ray | 185 A | A/B | 297-718 | PDB |
| 8T1J | EM | 270 A | A/B | 1-1429 | PDB |
| 8T1K | EM | 314 A | A/B | 1-1429 | PDB |
| AF-P29476-F1 | Predicted | AlphaFoldDB |
No variants for P29476
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P29476 | |||||
No associated diseases with P29476
7 regional properties for P29476
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 34 - 299 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 111 - 124 | IPR001245-1 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 151 - 169 | IPR001245-2 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 217 - 239 | IPR001245-3 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 263 - 285 | IPR001245-4 |
| active_site | Serine/threonine-protein kinase, active site | 157 - 169 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 40 - 63 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 1.14.13.39 | With NADH or NADPH as one donor, and incorporation of one atom of oxygen |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
30 GO annotations of cellular component
| Name | Definition |
|---|---|
| azurophil granule | Primary lysosomal granule found in neutrophil granulocytes. Contains a wide range of hydrolytic enzymes and is released into the extracellular fluid. |
| calyx of Held | The terminal specialization of a calyciferous axon which forms large synapses in the mammalian auditory central nervous system. |
| cell periphery | The part of a cell encompassing the cell cortex, the plasma membrane, and any external encapsulating structures. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nuclear membrane | Either of the lipid bilayers that surround the nucleus and form the nuclear envelope; excludes the intermembrane space. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| photoreceptor inner segment | The inner segment of a vertebrate photoreceptor containing mitochondria, ribosomes and membranes where opsin molecules are assembled and passed to be part of the outer segment discs. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| postsynaptic density, intracellular component | A network of proteins adjacent to the postsynaptic membrane forming an electron dense disc. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize neurotransmitter receptors in the adjacent membrane, such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| postsynaptic specialization, intracellular component | A network of proteins adjacent to the postsynaptic membrane. Its major components include the proteins that spatially and functionally organize neurotransmitter receptors in the adjacent membrane, such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| sarcolemma | The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers. |
| sarcoplasmic reticulum | A fine reticular network of membrane-limited elements that pervades the sarcoplasm of a muscle cell; continuous over large portions of the cell and with the nuclear envelope; that part of the endoplasmic reticulum specialized for calcium release, uptake and storage. |
| sarcoplasmic reticulum membrane | The lipid bilayer surrounding the sarcoplasmic reticulum. |
| secretory granule | A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
| T-tubule | Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane. |
| vesicle membrane | The lipid bilayer surrounding any membrane-bounded vesicle in the cell. |
| Z disc | Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached. |
18 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATPase binding | Binding to an ATPase, any enzyme that catalyzes the hydrolysis of ATP. |
| cadmium ion binding | Binding to a cadmium ion (Cd). |
| calcium-dependent protein binding | Binding to a protein or protein complex in the presence of calcium. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| flavin adenine dinucleotide binding | Binding to FAD, flavin-adenine dinucleotide, the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes, in either the oxidized form, FAD, or the reduced form, FADH2. |
| FMN binding | Binding to flavin mono nucleotide. Flavin mono nucleotide (FMN) is the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes. |
| heme binding | Binding to a heme, a compound composed of iron complexed in a porphyrin (tetrapyrrole) ring. |
| identical protein binding | Binding to an identical protein or proteins. |
| NADP binding | Binding to nicotinamide-adenine dinucleotide phosphate, a coenzyme involved in many redox and biosynthetic reactions; binding may be to either the oxidized form, NADP+, or the reduced form, NADPH. |
| NADPH binding | Binding to the reduced form, NADPH, of nicotinamide-adenine dinucleotide phosphate, a coenzyme involved in many redox and biosynthetic reactions. |
| nitric-oxide synthase activity | Catalysis of the reaction: L-arginine + n NADPH + n H+ + m O2 = citrulline + nitric oxide + n NADP+. |
| oxidoreductase activity | Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced. |
| phosphoprotein binding | Binding to a phosphorylated protein. |
| scaffold protein binding | Binding to a scaffold protein. Scaffold proteins are crucial regulators of many key signaling pathways. Although not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. |
| sodium channel regulator activity | Binds to and modulates the activity of a sodium channel. |
| transmembrane transporter binding | Binding to a transmembrane transporter, a protein or protein complex that enables the transfer of a substance, usually a specific substance or a group of related substances, from one side of a membrane to the other. |
| zinc ion binding | Binding to a zinc ion (Zn). |
63 GO annotations of biological process
| Name | Definition |
|---|---|
| aging | A developmental process that is a deterioration and loss of function over time. Aging includes loss of functions such as resistance to disease, homeostasis, and fertility, as well as wear and tear. Aging includes cellular senescence, but is more inclusive. May precede death and may succeed developmental maturation (GO:0021700). |
| arginine catabolic process | The chemical reactions and pathways resulting in the breakdown of arginine, 2-amino-5-(carbamimidamido)pentanoic acid. |
| behavioral response to cocaine | Any process that results in a change in the behavior of an organism as a result of a cocaine stimulus. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| cellular response to epinephrine stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an epinephrine stimulus. Epinephrine is a catecholamine that has the formula C9H13NO3; it is secreted by the adrenal medulla to act as a hormone, and released by certain neurons to act as a neurotransmitter active in the central nervous system. |
| cellular response to growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a growth factor stimulus. |
| cellular response to mechanical stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| establishment of protein localization | The directed movement of a protein to a specific location. |
| female pregnancy | The set of physiological processes that allow an embryo or foetus to develop within the body of a female animal. It covers the time from fertilization of a female ovum by a male spermatozoon until birth. |
| multicellular organismal response to stress | Any process that results in a change in state or activity of a multicellular organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation). |
| muscle contraction | A process in which force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of blood pressure | Any process in which the force of blood traveling through the circulatory system is decreased. |
| negative regulation of calcium ion transport | Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of calcium ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of cytosolic calcium ion concentration | Any process that decreases the concentration of calcium ions in the cytosol. |
| negative regulation of heart contraction | Any process that stops, prevents, or reduces the frequency, rate or extent of heart contraction. |
| negative regulation of hepatic stellate cell contraction | Any process that modulates stops, prevents, or reduces the frequency, rate or extent of hepatic stellate cell contraction. |
| negative regulation of hydrolase activity | Any process that stops or reduces the rate of hydrolase activity, the catalysis of the hydrolysis of various bonds. |
| negative regulation of insulin secretion | Any process that stops, prevents, or reduces the frequency, rate or extent of the regulated release of insulin. |
| negative regulation of iron ion transmembrane transport | Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of iron ions from one side of a membrane to the other by means of some agent such as a transporter or pore. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| negative regulation of peptidyl-serine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| negative regulation of potassium ion transport | Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of potassium ions (K+) into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| negative regulation of serotonin uptake | Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of serotonin into a cell. |
| negative regulation of vasoconstriction | Any process that stops, prevents, or reduces the frequency, rate or extent of vasoconstriction. |
| nitric oxide biosynthetic process | The chemical reactions and pathways resulting in the formation of nitric oxide, nitrogen monoxide (NO), a colorless gas only slightly soluble in water. |
| nitric oxide mediated signal transduction | Any intracellular signal transduction in which the signal is passed on within the cell via nitric oxide (NO). Includes synthesis of nitric oxide, receptors/sensors for nitric oxide (such as soluble guanylyl cyclase/sGC) and downstream effectors that further transmit the signal within the cell. Nitric oxide transmits its downstream effects through either cyclic GMP (cGMP)-dependent or independent mechanisms. |
| peptidyl-cysteine S-nitrosylation | The covalent addition of a nitric oxide (NO) group to the sulphur (S) atom of a cysteine residue in a protein, to form peptidyl-S-nitrosyl-L-cysteine. |
| positive regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | Any process that activates or increases the frequency, rate or extent of an adenylate cyclase-activating adrenergic receptor signaling pathway involved in some heart process. |
| positive regulation of guanylate cyclase activity | Any process that activates or increases the frequency, rate or extent of guanylate cyclase activity. |
| positive regulation of histone acetylation | Any process that activates or increases the frequency, rate or extent of the addition of an acetyl group to a histone protein. |
| positive regulation of long-term synaptic potentiation | Any process that activates or increases the frequency, rate or extent of long-term synaptic potentiation. |
| positive regulation of neuron death | Any process that activates or increases the frequency, rate or extent of neuron death. |
| positive regulation of peptidyl-serine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| positive regulation of sodium ion transmembrane transport | Any process that activates or increases the frequency, rate or extent of sodium ion transmembrane transport. |
| positive regulation of the force of heart contraction | Any process that increases the force of heart muscle contraction. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of heart contraction | Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process in which the heart decreases in volume in a characteristic way to propel blood through the body. |
| regulation of neurogenesis | Any process that modulates the frequency, rate or extent of neurogenesis, the generation of cells in the nervous system. |
| regulation of sensory perception of pain | Any process that modulates the frequency, rate or extent of the sensory perception of pain, the series of events required for an organism to receive a painful stimulus, convert it to a molecular signal, and recognize and characterize the signal. |
| regulation of sodium ion transport | Any process that modulates the frequency, rate or extent of the directed movement of sodium ions (Na+) into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| response to activity | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an activity stimulus. |
| response to estrogen | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of stimulus by an estrogen, C18 steroid hormones that can stimulate the development of female sexual characteristics. |
| response to ethanol | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ethanol stimulus. |
| response to heat | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a heat stimulus, a temperature stimulus above the optimal temperature for that organism. |
| response to hormone | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hormone stimulus. |
| response to hypoxia | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| response to lead ion | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lead ion stimulus. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| response to nicotine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nicotine stimulus. |
| response to nitric oxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nitric oxide stimulus. |
| response to nutrient levels | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients. |
| response to organic cyclic compound | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic cyclic compound stimulus. |
| response to organonitrogen compound | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organonitrogen stimulus. An organonitrogen compound is formally a compound containing at least one carbon-nitrogen bond. |
| response to peptide hormone | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptide hormone stimulus. A peptide hormone is any of a class of peptides that are secreted into the blood stream and have endocrine functions in living animals. |
| response to vitamin E | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin E stimulus. |
| retrograde trans-synaptic signaling by nitric oxide | Cell-cell signaling from postsynapse to presynapse, across the synaptic cleft, mediated by nitric oxide. |
| striated muscle contraction | A process in which force is generated within striated muscle tissue, resulting in the shortening of the muscle. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. Striated muscle is a type of muscle in which the repeating units (sarcomeres) of the contractile myofibrils are arranged in registry throughout the cell, resulting in transverse or oblique striations observable at the level of the light microscope. |
| synaptic signaling by nitric oxide | Cell-cell signaling to or from a synapse, mediated by nitric oxide. |
| vasodilation | An increase in the internal diameter of blood vessels, especially arterioles or capillaries, due to relaxation of smooth muscle cells that line the vessels, and usually resulting in a decrease in blood pressure. |
| xenobiotic catabolic process | The chemical reactions and pathways resulting in the breakdown of a xenobiotic compound, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P29473 | NOS3 | Nitric oxide synthase, endothelial | Bos taurus (Bovine) | EV |
| Q9UBK8 | MTRR | Methionine synthase reductase | Homo sapiens (Human) | PR |
| P29474 | NOS3 | Nitric oxide synthase, endothelial | Homo sapiens (Human) | SS |
| P29475 | NOS1 | Nitric oxide synthase, brain | Homo sapiens (Human) | SS |
| P70313 | Nos3 | Nitric oxide synthase 3 | Mus musculus (Mouse) | SS |
| Q8C1A3 | Mtrr | Methionine synthase reductase | Mus musculus (Mouse) | PR |
| Q28969 | NOS3 | Nitric oxide synthase 3 | Sus scrofa (Pig) | SS |
| Q498R1 | Mtrr | Methionine synthase reductase | Rattus norvegicus (Rat) | PR |
| Q62600 | Nos3 | Nitric oxide synthase 3 | Rattus norvegicus (Rat) | SS |
| Q9FKW6 | LFNR1 | Ferredoxin--NADP reductase, leaf isozyme 1, chloroplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEENTFGVQQ | IQPNVISVRL | FKRKVGGLGF | LVKERVSKPP | VIISDLIRGG | AAEQSGLIQA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GDIILAVNDR | PLVDLSYDSA | LEVLRGIASE | THVVLILRGP | EGFTTHLETT | FTGDGTPKTI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RVTQPLGPPT | KAVDLSHQPS | ASKDQSLAVD | RVTGLGNGPQ | HAQGHGQGAG | SVSQANGVAI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DPTMKSTKAN | LQDIGEHDEL | LKEIEPVLSI | LNSGSKATNR | GGPAKAEMKD | TGIQVDRDLD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GKSHKAPPLG | GDNDRVFNDL | WGKDNVPVIL | NNPYSEKEQS | PTSGKQSPTK | NGSPSRCPRF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LKVKNWETDV | VLTDTLHLKS | TLETGCTEHI | CMGSIMLPSQ | HTRKPEDVRT | KDQLFPLAKE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FLDQYYSSIK | RFGSKAHMDR | LEEVNKEIES | TSTYQLKDTE | LIYGAKHAWR | NASRCVGRIQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| WSKLQVFDAR | DCTTAHGMFN | YICNHVKYAT | NKGNLRSAIT | IFPQRTDGKH | DFRVWNSQLI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RYAGYKQPDG | STLGDPANVQ | FTEICIQQGW | KAPRGRFDVL | PLLLQANGND | PELFQIPPEL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VLEVPIRHPK | FDWFKDLGLK | WYGLPAVSNM | LLEIGGLEFS | ACPFSGWYMG | TEIGVRDYCD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| NSRYNILEEV | AKKMDLDMRK | TSSLWKDQAL | VEINIAVLYS | FQSDKVTIVD | HHSATESFIK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HMENEYRCRG | GCPADWVWIV | PPMSGSITPV | FHQEMLNYRL | TPSFEYQPDP | WNTHVWKGTN |
| 730 | 740 | 750 | 760 | 770 | 780 |
| GTPTKRRAIG | FKKLAEAVKF | SAKLMGQAMA | KRVKATILYA | TETGKSQAYA | KTLCEIFKHA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FDAKAMSMEE | YDIVHLEHEA | LVLVVTSTFG | NGDPPENGEK | FGCALMEMRH | PNSVQEERKS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| YKVRFNSVSS | YSDSRKSSGD | GPDLRDNFES | TGPLANVRFS | VFGLGSRAYP | HFCAFGHAVD |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TLLEELGGER | ILKMREGDEL | CGQEEAFRTW | AKKVFKAACD | VFCVGDDVNI | EKPNNSLISN |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| DRSWKRNKFR | LTYVAEAPDL | TQGLSNVHKK | RVSAARLLSR | QNLQSPKFSR | STIFVRLHTN |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| GNQELQYQPG | DHLGVFPGNH | EDLVNALIER | LEDAPPANHV | VKVEMLEERN | TALGVISNWK |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| DESRLPPCTI | FQAFKYYLDI | TTPPTPLQLQ | QFASLATNEK | EKQRLLVLSK | GLQEYEEWKW |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| GKNPTMVEVL | EEFPSIQMPA | TLLLTQLSLL | QPRYYSISSS | PDMYPDEVHL | TVAIVSYHTR |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| DGEGPVHHGV | CSSWLNRIQA | DDVVPCFVRG | APSFHLPRNP | QVPCILVGPG | TGIAPFRSFW |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| QQRQFDIQHK | GMNPCPMVLV | FGCRQSKIDH | IYREETLQAK | NKGVFRELYT | AYSREPDRPK |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| KYVQDVLQEQ | LAESVYRALK | EQGGHIYVCG | DVTMAADVLK | AIQRIMTQQG | KLSEEDAGVF |
| 1390 | 1400 | 1410 | 1420 | ||
| ISRLRDDNRY | HEDIFGVTLR | TYEVTNRLRS | ESIAFIEESK | KDADEVFSS |