P29473
Gene name |
NOS3 |
Protein name |
Nitric oxide synthase, endothelial |
Names |
Constitutive NOS, cNOS, EC-NOS, Endothelial NOS, eNOS, NOS type III, NOSIII |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
|
EC number |
|
Protein Class |
NITRIC OXIDE SYNTHASE OXYGENASE (PTHR43410) |
Descriptions
Nitric oxide synthase produces nitric oxide (NO) which is implicated in vascular smooth muscle relaxation through a cGMP-mediated signal transduction pathway. The primary sequences of the three mammalian nitric oxide synthase (NOS) isoforms differ by the insertion of a 52-55-amino acid loop into the reductase domains of the endothelial (eNOS) and neuronal (nNOS), but not inducible (iNOS). The loop mediates the autoinhibition in eNOS and nNOS. Deletion of the autoinhibitory loop increases the catalytic activity of eNOS.
Autoinhibitory domains (AIDs)
Target domain |
122-483 (NO synthase domain); 732-1164 (FAD/NAD-binding domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay |
Accessory elements
No accessory elements
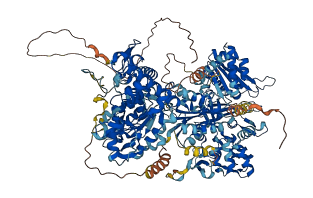
Autoinhibited structure

Activated structure

129 structures for P29473
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1D0C | X-ray | 165 A | A/B | 39-482 | PDB |
| 1D0O | X-ray | 195 A | A/B | 39-482 | PDB |
| 1D1V | X-ray | 193 A | A/B | 39-482 | PDB |
| 1D1W | X-ray | 200 A | A/B | 39-482 | PDB |
| 1D1X | X-ray | 200 A | A/B | 39-482 | PDB |
| 1D1Y | X-ray | 220 A | A/B | 39-482 | PDB |
| 1DM6 | X-ray | 195 A | A/B | 39-482 | PDB |
| 1DM7 | X-ray | 210 A | A/B | 39-482 | PDB |
| 1DM8 | X-ray | 225 A | A/B | 39-482 | PDB |
| 1DMI | X-ray | 200 A | A/B | 39-482 | PDB |
| 1DMJ | X-ray | 235 A | A/B | 39-482 | PDB |
| 1DMK | X-ray | 190 A | A/B | 39-482 | PDB |
| 1ED4 | X-ray | 186 A | A/B | 39-482 | PDB |
| 1ED5 | X-ray | 180 A | A/B | 39-482 | PDB |
| 1ED6 | X-ray | 205 A | A/B | 39-482 | PDB |
| 1FOI | X-ray | 193 A | A/B | 39-482 | PDB |
| 1FOJ | X-ray | 210 A | A/B | 39-482 | PDB |
| 1FOL | X-ray | 220 A | A/B | 39-482 | PDB |
| 1FOO | X-ray | 200 A | A/B | 39-482 | PDB |
| 1FOP | X-ray | 230 A | A/B | 39-482 | PDB |
| 1I83 | X-ray | 200 A | A/B | 39-482 | PDB |
| 1NSE | X-ray | 190 A | A/B | 39-482 | PDB |
| 1P6L | X-ray | 235 A | A/B | 67-483 | PDB |
| 1P6M | X-ray | 227 A | A/B | 67-483 | PDB |
| 1P6N | X-ray | 250 A | A/B | 67-483 | PDB |
| 1Q2O | X-ray | 174 A | A/B | 67-482 | PDB |
| 1RS8 | X-ray | 230 A | A/B | 67-482 | PDB |
| 1RS9 | X-ray | 222 A | A/B | 67-482 | PDB |
| 1ZZS | X-ray | 185 A | A/B | 67-482 | PDB |
| 1ZZT | X-ray | 214 A | A/B | 67-482 | PDB |
| 2G6O | X-ray | 190 A | A/B | 67-482 | PDB |
| 2HX2 | X-ray | 195 A | A/B | 67-482 | PDB |
| 2NSE | X-ray | 234 A | A/B | 39-482 | PDB |
| 3DQS | X-ray | 203 A | A/B | 67-482 | PDB |
| 3DQT | X-ray | 254 A | A/B | 67-482 | PDB |
| 3E7S | X-ray | 250 A | A/B | 57-487 | PDB |
| 3JWW | X-ray | 220 A | A/B | 39-482 | PDB |
| 3JWX | X-ray | 200 A | A/B | 39-482 | PDB |
| 3JWY | X-ray | 224 A | A/B | 39-482 | PDB |
| 3JWZ | X-ray | 240 A | A/B | 39-482 | PDB |
| 3N5P | X-ray | 239 A | A/B | 39-482 | PDB |
| 3N5Q | X-ray | 290 A | A/B | 39-482 | PDB |
| 3N5R | X-ray | 257 A | A/B | 39-482 | PDB |
| 3N5S | X-ray | 218 A | A/B | 39-482 | PDB |
| 3N5T | X-ray | 252 A | A/B | 39-482 | PDB |
| 3NLD | X-ray | 228 A | A/B | 39-482 | PDB |
| 3NLE | X-ray | 195 A | A/B | 39-482 | PDB |
| 3NLF | X-ray | 232 A | A/B | 39-482 | PDB |
| 3NLG | X-ray | 238 A | A/B | 39-482 | PDB |
| 3NLH | X-ray | 210 A | A/B | 39-482 | PDB |
| 3NLI | X-ray | 198 A | A/B | 39-482 | PDB |
| 3NLT | X-ray | 274 A | A/B | 39-482 | PDB |
| 3NLU | X-ray | 265 A | A/B | 39-482 | PDB |
| 3NSE | X-ray | 210 A | A/B | 39-482 | PDB |
| 3PNH | X-ray | 193 A | A/B | 67-482 | PDB |
| 3RQO | X-ray | 208 A | A/B | 40-482 | PDB |
| 3RQP | X-ray | 235 A | A/B | 40-482 | PDB |
| 4C3A | X-ray | 220 A | A/B | 40-482 | PDB |
| 4CAR | X-ray | 205 A | A/B | 40-482 | PDB |
| 4CFT | X-ray | 179 A | A/B | 40-482 | PDB |
| 4CTY | X-ray | 230 A | A/B | 40-482 | PDB |
| 4CTZ | X-ray | 201 A | A/B | 40-482 | PDB |
| 4CU0 | X-ray | 208 A | A/B | 40-482 | PDB |
| 4CU1 | X-ray | 189 A | A/B | 40-482 | PDB |
| 4CUL | X-ray | 223 A | A/B | 40-482 | PDB |
| 4CUM | X-ray | 233 A | A/B | 40-482 | PDB |
| 4CUN | X-ray | 248 A | A/B | 40-482 | PDB |
| 4CVG | X-ray | 231 A | A/B | 40-482 | PDB |
| 4CWV | X-ray | 234 A | A/B | 40-482 | PDB |
| 4CWW | X-ray | 216 A | A/B | 40-482 | PDB |
| 4CWX | X-ray | 215 A | A/B | 40-482 | PDB |
| 4CWY | X-ray | 215 A | A/B | 40-482 | PDB |
| 4CWZ | X-ray | 208 A | A/B | 40-482 | PDB |
| 4CX0 | X-ray | 220 A | A/B | 40-482 | PDB |
| 4CX1 | X-ray | 213 A | A/B | 40-482 | PDB |
| 4CX2 | X-ray | 204 A | A/B | 40-482 | PDB |
| 4D33 | X-ray | 209 A | A/B | 40-482 | PDB |
| 4D34 | X-ray | 225 A | A/B | 40-482 | PDB |
| 4D35 | X-ray | 218 A | A/B | 40-482 | PDB |
| 4D36 | X-ray | 205 A | A/B | 40-482 | PDB |
| 4D37 | X-ray | 210 A | A/B | 40-482 | PDB |
| 4D38 | X-ray | 230 A | A/B | 40-482 | PDB |
| 4D39 | X-ray | 200 A | A/B | 40-482 | PDB |
| 4D3A | X-ray | 225 A | A/B | 40-482 | PDB |
| 4IMX | X-ray | 225 A | A/B | 40-482 | PDB |
| 4JSK | X-ray | 228 A | A/B | 40-482 | PDB |
| 4JSL | X-ray | 204 A | A/B | 40-482 | PDB |
| 4JSM | X-ray | 225 A | A/B | 40-482 | PDB |
| 4K5H | X-ray | 225 A | A/B | 40-482 | PDB |
| 4K5I | X-ray | 208 A | A/B | 40-482 | PDB |
| 4K5J | X-ray | 236 A | A/B | 40-482 | PDB |
| 4K5K | X-ray | 200 A | A/B | 40-482 | PDB |
| 4KCP | X-ray | 207 A | A/B | 40-482 | PDB |
| 4KCQ | X-ray | 203 A | A/B | 40-482 | PDB |
| 4KCR | X-ray | 209 A | A/B | 40-482 | PDB |
| 4KCS | X-ray | 205 A | A/B | 40-482 | PDB |
| 4LUW | X-ray | 225 A | A/B | 41-482 | PDB |
| 4NSE | X-ray | 195 A | A/B | 39-482 | PDB |
| 4UH7 | X-ray | 223 A | A/B | 40-482 | PDB |
| 4UH8 | X-ray | 230 A | A/B | 40-482 | PDB |
| 4UH9 | X-ray | 214 A | A/B | 40-482 | PDB |
| 4UHA | X-ray | 220 A | A/B | 40-482 | PDB |
| 4UPQ | X-ray | 203 A | A/B | 40-482 | PDB |
| 4UPR | X-ray | 193 A | A/B | 40-482 | PDB |
| 4UPS | X-ray | 195 A | A/B | 40-482 | PDB |
| 4UPT | X-ray | 220 A | A/B | 40-482 | PDB |
| 5ADJ | X-ray | 222 A | A/B | 40-482 | PDB |
| 5ADK | X-ray | 180 A | A/B | 40-482 | PDB |
| 5ADL | X-ray | 221 A | A/B | 40-482 | PDB |
| 5ADM | X-ray | 220 A | A/B | 40-482 | PDB |
| 5ADN | X-ray | 200 A | A/B | 40-482 | PDB |
| 5FJ2 | X-ray | 205 A | A/B | 40-482 | PDB |
| 5FJ3 | X-ray | 220 A | A/B | 40-482 | PDB |
| 5FVY | X-ray | 210 A | A/B | 40-482 | PDB |
| 5FVZ | X-ray | 205 A | A/B | 40-482 | PDB |
| 5NSE | X-ray | 190 A | A/B | 39-482 | PDB |
| 5UOD | X-ray | 201 A | A/B | 40-482 | PDB |
| 5VV6 | X-ray | 200 A | A/B | 40-482 | PDB |
| 5VV7 | X-ray | 220 A | A/B | 40-482 | PDB |
| 5VV8 | X-ray | 215 A | A/B | 40-482 | PDB |
| 5VV9 | X-ray | 250 A | A/B | 40-482 | PDB |
| 5VVA | X-ray | 255 A | A/B | 40-482 | PDB |
| 5VVG | X-ray | 230 A | A/B | 40-482 | PDB |
| 5VVN | X-ray | 240 A | A/B | 40-482 | PDB |
| 6NSE | X-ray | 235 A | A/B | 39-482 | PDB |
| 7NSE | X-ray | 235 A | A/B | 39-482 | PDB |
| 8NSE | X-ray | 225 A | A/B | 39-482 | PDB |
| 9NSE | X-ray | 224 A | A/B | 39-482 | PDB |
| AF-P29473-F1 | Predicted | AlphaFoldDB |
No variants for P29473
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P29473 | |||||
No associated diseases with P29473
17 regional properties for P29473
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Flavodoxin-like | 523 - 536 | IPR001094-1 |
| domain | Flavodoxin-like | 570 - 581 | IPR001094-2 |
| domain | Flavodoxin-like | 646 - 656 | IPR001094-3 |
| domain | Flavodoxin-like | 670 - 689 | IPR001094-4 |
| domain | Oxidoreductase FAD/NAD(P)-binding | 1013 - 1125 | IPR001433 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 795 - 805 | IPR001709-1 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 940 - 947 | IPR001709-2 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 976 - 985 | IPR001709-3 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 1012 - 1031 | IPR001709-4 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 1042 - 1051 | IPR001709-5 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 1055 - 1066 | IPR001709-6 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 1087 - 1103 | IPR001709-7 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 1112 - 1120 | IPR001709-8 |
| domain | Sulfite reductase [NADPH] flavoprotein alpha-component-like, FAD-binding | 754 - 981 | IPR003097 |
| domain | Nitric oxide synthase, N-terminal | 122 - 483 | IPR004030 |
| domain | Flavodoxin/nitric oxide synthase | 522 - 705 | IPR008254 |
| domain | FAD-binding domain, ferredoxin reductase-type | 758 - 1004 | IPR017927 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR43410 | NITRIC OXIDE SYNTHASE OXYGENASE |
| PANTHER Subfamily | PTHR43410:SF1 | NITRIC OXIDE SYNTHASE OXYGENASE |
| PANTHER Protein Class | protein modifying enzyme | |
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| caveola | A membrane raft that forms small pit, depression, or invagination that communicates with the outside of a cell and extends inward, indenting the cytoplasm and the cell membrane. Examples include flask-shaped invaginations of the plasma membrane in adipocytes associated with caveolin proteins, and minute pits or incuppings of the cell membrane formed during pinocytosis. Caveolae may be pinched off to form free vesicles within the cytoplasm. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| flavin adenine dinucleotide binding | Binding to FAD, flavin-adenine dinucleotide, the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes, in either the oxidized form, FAD, or the reduced form, FADH2. |
| FMN binding | Binding to flavin mono nucleotide. Flavin mono nucleotide (FMN) is the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes. |
| heme binding | Binding to a heme, a compound composed of iron complexed in a porphyrin (tetrapyrrole) ring. |
| metal ion binding | Binding to a metal ion. |
| NADP binding | Binding to nicotinamide-adenine dinucleotide phosphate, a coenzyme involved in many redox and biosynthetic reactions; binding may be to either the oxidized form, NADP+, or the reduced form, NADPH. |
| nitric-oxide synthase activity | Catalysis of the reaction: L-arginine + n NADPH + n H+ + m O2 = citrulline + nitric oxide + n NADP+. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| arginine catabolic process | The chemical reactions and pathways resulting in the breakdown of arginine, 2-amino-5-(carbamimidamido)pentanoic acid. |
| blood coagulation | The sequential process in which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers. |
| cellular response to laminar fluid shear stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a laminar fluid shear stress stimulus. Laminar fluid flow is the force acting on an object in a system where the fluid is moving across a solid surface in parallel layers. |
| mitochondrion organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a mitochondrion; includes mitochondrial morphogenesis and distribution, and replication of the mitochondrial genome as well as synthesis of new mitochondrial components. |
| negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway via death domain receptors. |
| negative regulation of leukocyte cell-cell adhesion | Any process that stops, prevents or reduces the frequency, rate or extent of leukocyte cell-cell adhesion. |
| nitric oxide biosynthetic process | The chemical reactions and pathways resulting in the formation of nitric oxide, nitrogen monoxide (NO), a colorless gas only slightly soluble in water. |
| positive regulation of guanylate cyclase activity | Any process that activates or increases the frequency, rate or extent of guanylate cyclase activity. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1JPJ0 | NDOR1 | NADPH-dependent diflavin oxidoreductase 1 | Bos taurus (Bovine) | PR |
| Q9TUX8 | NOS3 | Nitric oxide synthase 3 | Canis lupus familiaris (Dog) (Canis familiaris) | SS |
| Q9UBK8 | MTRR | Methionine synthase reductase | Homo sapiens (Human) | PR |
| P29475 | NOS1 | Nitric oxide synthase, brain | Homo sapiens (Human) | SS |
| P29474 | NOS3 | Nitric oxide synthase, endothelial | Homo sapiens (Human) | SS |
| Q8C1A3 | Mtrr | Methionine synthase reductase | Mus musculus (Mouse) | PR |
| P70313 | Nos3 | Nitric oxide synthase 3 | Mus musculus (Mouse) | SS |
| Q28969 | NOS3 | Nitric oxide synthase 3 | Sus scrofa (Pig) | SS |
| Q498R1 | Mtrr | Methionine synthase reductase | Rattus norvegicus (Rat) | PR |
| P29476 | Nos1 | Nitric oxide synthase, brain | Rattus norvegicus (Rat) | SS |
| Q62600 | Nos3 | Nitric oxide synthase 3 | Rattus norvegicus (Rat) | SS |
| Q9FKW6 | LFNR1 | Ferredoxin--NADP reductase, leaf isozyme 1, chloroplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGNLKSVGQE | PGPPCGLGLG | LGLGLCGKQG | PASPAPEPSR | APAPATPHAP | DHSPAPNSPT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LTRPPEGPKF | PRVKNWELGS | ITYDTLCAQS | QQDGPCTPRC | CLGSLVLPRK | LQTRPSPGPP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PAEQLLSQAR | DFINQYYSSI | KRSGSQAHEE | RLQEVEAEVA | STGTYHLRES | ELVFGAKQAW |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RNAPRCVGRI | QWGKLQVFDA | RDCSSAQEMF | TYICNHIKYA | TNRGNLRSAI | TVFPQRAPGR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GDFRIWNSQL | VRYAGYRQQD | GSVRGDPANV | EITELCIQHG | WTPGNGRFDV | LPLLLQAPDE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| APELFVLPPE | LVLEVPLEHP | TLEWFAALGL | RWYALPAVSN | MLLEIGGLEF | SAAPFSGWYM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| STEIGTRNLC | DPHRYNILED | VAVCMDLDTR | TTSSLWKDKA | AVEINLAVLH | SFQLAKVTIV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DHHAATVSFM | KHLDNEQKAR | GGCPADWAWI | VPPISGSLTP | VFHQEMVNYI | LSPAFRYQPD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PWKGSATKGA | GITRKKTFKE | VANAVKISAS | LMGTLMAKRV | KATILYASET | GRAQSYAQQL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GRLFRKAFDP | RVLCMDEYDV | VSLEHEALVL | VVTSTFGNGD | PPENGESFAA | ALMEMSGPYN |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SSPRPEQHKS | YKIRFNSVSC | SDPLVSSWRR | KRKESSNTDS | AGALGTLRFC | VFGLGSRAYP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HFCAFARAVD | TRLEELGGER | LLQLGQGDEL | CGQEEAFRGW | AKAAFQASCE | TFCVGEEAKA |
| 730 | 740 | 750 | 760 | 770 | 780 |
| AAQDIFSPKR | SWKRQRYRLS | TQAEGLQLLP | GLIHVHRRKM | FQATVLSVEN | LQSSKSTRAT |
| 790 | 800 | 810 | 820 | 830 | 840 |
| ILVRLDTAGQ | EGLQYQPGDH | IGICPPNRPG | LVEALLSRVE | DPPPPTESVA | VEQLEKGSPG |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GPPPSWVRDP | RLPPCTLRQA | LTFFLDITSP | PSPRLLRLLS | TLAEEPSEQQ | ELETLSQDPR |
| 910 | 920 | 930 | 940 | 950 | 960 |
| RYEEWKWFRC | PTLLEVLEQF | PSVALPAPLL | LTQLPLLQPR | YYSVSSAPNA | HPGEVHLTVA |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| VLAYRTQDGL | GPLHYGVCST | WLSQLKTGDP | VPCFIRGAPS | FRLPPDPYVP | CILVGPGTGI |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| APFRGFWQER | LHDIESKGLQ | PAPMTLVFGC | RCSQLDHLYR | DEVQDAQERG | VFGRVLTAFS |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| REPDSPKTYV | QDILRTELAA | EVHRVLCLER | GHMFVCGDVT | MATSVLQTVQ | RILATEGDME |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| LDEAGDVIGV | LRDQQRYHED | IFGLTLRTQE | VTSRIRTQSF | SLQERHLRGA | VPWAFDPPGP |
| DTPGP |