P29350
Gene name |
PTPN6 (HCP, PTP1C) |
Protein name |
Tyrosine-protein phosphatase non-receptor type 6 |
Names |
Hematopoietic cell protein-tyrosine phosphatase, Protein-tyrosine phosphatase 1C, PTP-1C, Protein-tyrosine phosphatase SHP-1, SH-PTP1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5777 |
EC number |
3.1.3.48: Phosphoric monoester hydrolases |
Protein Class |
TYROSINE-PROTEIN PHOSPHATASE CORKSCREW (PTHR46257) |
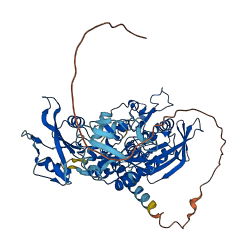
Descriptions
PTPN6 is a Src homology 2 (SH2) domain-containing protein tyrosine phosphatase (PTPase). Its N-terminal SH2 domain autoinhibits the function of the kinase domain via intramolecular interaction. The N-SH2 domain blocks the active site of the PTPase domain (catalytic domain) in the closed conformation. Ligand binding to SH2 domains will cause conformational changes, resulting in the release of the N-SH2 domain from the PTPase domain. This will trigger the rotation of the C-SH2 domain, the repositioning of the N-SH2 domain, and consequent exposure of the active site of the catalytic domain.
Autoinhibitory domains (AIDs)
Target domain |
253-518 (catalytic domain of tyrosine-protein phosphatase non-receptor type 6) |
Relief mechanism |
Ligand binding |
Assay |
Deletion assay, Structural analysis |
Accessory elements
No accessory elements
References
- Pei D et al. (1994) "Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: a new function for Src homology 2 domains", Biochemistry, 33, 15483-93
- Wang W et al. (2011) "Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation", Journal of cellular biochemistry, 112, 2062-71
Autoinhibited structure
Activated structure

14 structures for P29350
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1FPR | X-ray | 250 A | A | 243-526 | PDB |
| 1GWZ | X-ray | 250 A | A | 243-541 | PDB |
| 1X6C | NMR | - | A | 110-214 | PDB |
| 2B3O | X-ray | 280 A | A | 1-532 | PDB |
| 2RMX | NMR | - | A | 110-214 | PDB |
| 2YU7 | NMR | - | A | 110-214 | PDB |
| 3PS5 | X-ray | 310 A | A | 1-595 | PDB |
| 4GRY | X-ray | 170 A | A | 243-528 | PDB |
| 4GRZ | X-ray | 137 A | A | 243-528 | PDB |
| 4GS0 | X-ray | 180 A | A/B | 243-528 | PDB |
| 4HJP | X-ray | 140 A | A | 243-528 | PDB |
| 4HJQ | X-ray | 180 A | A/B | 243-528 | PDB |
| 6SM5 | X-ray | 275 A | A | 100-214 | PDB |
| AF-P29350-F1 | Predicted | AlphaFoldDB |
323 variants for P29350
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1591683472 CA383747263 |
2 | V>G | No |
ClinGen Ensembl |
|
|
CA383747329 rs1555147752 |
4 | W>C | No |
ClinGen gnomAD |
|
|
COSM3739862 CA232442152 COSM3739863 rs769682837 |
7 | R>Q | liver [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1555147756 CA383747424 |
12 | L>R | No |
ClinGen gnomAD |
|
|
CA6421460 rs782158849 |
15 | E>K | No |
ClinGen ExAC |
|
|
rs782702822 CA6421461 |
16 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA383747470 rs1555147762 |
17 | L>V | No |
ClinGen gnomAD |
|
|
CA383747487 rs1555147765 |
19 | K>E | No |
ClinGen gnomAD |
|
|
CA6421463 rs377003393 |
20 | G>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs11547853 CA232442165 |
28 | L>P | No |
ClinGen Ensembl |
|
|
CA6421466 rs782523221 |
33 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs369614203 CA6421467 |
33 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs369614203 CA383747623 |
33 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1555147772 CA383747624 |
34 | K>Q | No |
ClinGen gnomAD |
|
|
rs782452891 CA6421469 |
38 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782042492 CA6421470 |
40 | S>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs267603654 CA232442178 |
42 | S>F | No |
ClinGen Ensembl |
|
|
rs377254990 CA6421472 |
43 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1555147854 CA383747789 |
45 | V>M | No |
ClinGen gnomAD |
|
|
rs782712760 CA6421508 |
46 | G>W | No |
ClinGen ExAC gnomAD |
|
|
rs781820797 CA6421509 |
47 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555147857 CA383747878 |
53 | R>Q | No |
ClinGen gnomAD |
|
|
rs370939978 COSM179634 CA6421510 |
53 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1555147859 CA383747896 |
55 | Q>* | No |
ClinGen gnomAD |
|
|
CA6421511 rs782618719 |
59 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA6421513 rs782531361 |
70 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA6421515 rs782204971 |
73 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs782242482 CA6421518 |
80 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA6421519 rs782393921 |
83 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1048597 CA232442300 |
86 | L>V | No |
ClinGen Ensembl |
|
|
CA6421522 COSM3384601 COSM3384600 rs782163170 |
89 | R>C | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs782308449 CA6421523 |
89 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs782302155 CA6421524 |
90 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3384603 COSM3384602 rs1555147881 CA383748279 |
91 | G>D | pancreas [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1342222997 CA383748295 |
93 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA232442313 rs1028611737 |
99 | P>A | No |
ClinGen Ensembl |
|
|
CA383748370 rs1555147885 |
99 | P>L | No |
ClinGen gnomAD |
|
|
CA383748400 rs1555147887 |
102 | C>S | No |
ClinGen gnomAD |
|
|
rs782804780 CA6421529 |
107 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA383748669 rs1189982819 |
115 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA6421558 rs782645298 |
118 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1555148312 CA383748713 |
122 | T>A | No |
ClinGen gnomAD |
|
|
rs201550362 CA6421559 |
122 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6421563 rs782364925 |
127 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383748748 rs1555148318 |
128 | G>S | No |
ClinGen gnomAD |
|
|
CA6421566 COSM1242731 rs782797143 |
132 | T>M | oesophagus [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs782054048 CA6421568 |
135 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs782710028 CA6421569 |
136 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1228369426 CA383748803 |
136 | R>H | No |
ClinGen TOPMed |
|
|
CA6421570 rs781818617 |
139 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA232443273 rs1048598 |
146 | V>E | No |
ClinGen Ensembl |
|
|
rs782775329 CA6421572 |
153 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs781874476 CA6421573 |
154 | P>H | No |
ClinGen ExAC gnomAD |
|
|
rs782700653 CA6421574 |
157 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs782700653 CA6421575 |
157 | G>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA6421577 rs373750986 |
161 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1555148329 CA383748973 |
161 | P>S | No |
ClinGen gnomAD |
|
|
CA6421580 rs782394570 |
163 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs369530834 CA6421581 |
163 | R>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1555148338 CA383749010 |
166 | H>Y | No |
ClinGen gnomAD |
|
|
CA6421582 rs782299688 |
170 | M>T | No |
ClinGen ExAC |
|
|
rs1555148341 CA383749057 |
171 | C>Y | No |
ClinGen gnomAD |
|
|
CA6421584 rs781941174 |
172 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1555148370 CA383749081 |
173 | G>S | No |
ClinGen gnomAD |
|
|
CA383749089 rs1555148372 |
174 | G>R | No |
ClinGen gnomAD |
|
|
CA232443452 rs781988918 |
175 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA383749102 rs1281119726 |
175 | R>H | No |
ClinGen TOPMed |
|
|
rs970406672 CA232443456 |
182 | E>K | No |
ClinGen Ensembl |
|
|
CA383749167 rs782304451 |
183 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421610 rs782304451 |
183 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383749179 rs1565581803 |
184 | F>S | No |
ClinGen Ensembl |
|
|
CA383749185 rs1591687142 |
185 | D>A | No |
ClinGen Ensembl |
|
|
rs782079530 CA383749184 |
185 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs782079530 CA6421612 COSM943094 |
185 | D>N | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA383749207 rs368426352 |
188 | T>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs368426352 CA6421613 |
188 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781875658 CA383749215 |
189 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA6421614 rs781875658 |
189 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA383749209 rs1555148380 |
189 | D>N | No |
ClinGen Ensembl |
|
|
rs1555148382 CA383749217 |
190 | L>V | No |
ClinGen gnomAD |
|
|
CA383749237 rs1555148387 |
193 | H>Y | No |
ClinGen gnomAD |
|
|
rs782576858 CA6421616 |
197 | T>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA383749294 rs1555148393 |
201 | E>Q | No |
ClinGen gnomAD |
|
|
CA383749316 rs1555148396 |
204 | G>D | No |
ClinGen gnomAD |
|
|
rs1555148401 CA383749320 |
205 | A>D | No |
ClinGen gnomAD |
|
|
CA6421620 rs375296528 |
205 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs375296528 CA6421619 |
205 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA383749322 rs1555148401 |
205 | A>V | No |
ClinGen gnomAD |
|
|
CA383749330 rs1469972531 |
206 | F>L | No |
ClinGen TOPMed |
|
|
CA383749324 rs1555148406 |
206 | F>L | No |
ClinGen gnomAD |
|
|
rs1159545254 CA383749326 |
206 | F>Y | No |
ClinGen TOPMed |
|
|
CA6421622 rs367551491 COSM179638 |
210 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs973281293 CA232443499 |
210 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA383749377 rs1555148454 |
212 | P>L | No |
ClinGen gnomAD |
|
|
rs1555148455 CA383749387 |
214 | Y>H | No |
ClinGen gnomAD |
|
|
rs1591687479 CA383749415 |
218 | V>G | No |
ClinGen Ensembl |
|
|
CA6421644 rs782524619 |
218 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA383749429 rs1212719021 |
220 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA6421646 rs782302331 |
223 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA6421647 rs201345298 |
225 | N>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs781935337 CA383749469 |
226 | R>P | No |
ClinGen ExAC gnomAD |
|
|
CA6421648 rs781935337 |
226 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs782212727 CA6421649 |
231 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA383749526 rs1340881647 |
234 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA383749538 rs1270207675 |
236 | S>A | No |
ClinGen TOPMed gnomAD |
|
|
CA6421650 rs782356549 |
236 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1315244865 CA383749545 |
237 | E>D | No |
ClinGen TOPMed |
|
|
CA6421652 rs782130684 |
237 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421653 rs782130684 |
237 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383749549 rs1555148481 |
238 | D>Y | No |
ClinGen gnomAD |
|
|
CA6421655 rs782045447 |
240 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1555148485 CA383749570 |
241 | K>R | No |
ClinGen gnomAD |
|
|
rs1555148488 CA383749586 |
244 | F>I | No |
ClinGen gnomAD |
|
|
CA383749600 rs1555148490 |
245 | W>* | No |
ClinGen gnomAD |
|
|
rs781816472 CA6421657 |
246 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA6421678 rs782819686 |
259 | L>S | No |
ClinGen ExAC gnomAD |
|
|
rs1591687896 CA383749786 |
260 | H>P | No |
ClinGen Ensembl |
|
|
CA383749790 rs1555148535 |
260 | H>Q | No |
ClinGen gnomAD |
|
|
CA383749792 rs1555148535 |
260 | H>Q | No |
ClinGen gnomAD |
|
|
rs1555148529 CA383749785 |
260 | H>Y | No |
ClinGen gnomAD |
|
|
rs761631984 CA232443808 |
262 | R>H | No |
ClinGen gnomAD |
|
|
rs781848123 CA383749863 |
267 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781848123 CA6421682 |
267 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383749861 rs781848123 |
267 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555148541 CA383749860 |
267 | R>W | No |
ClinGen gnomAD |
|
|
CA6421684 rs782637799 |
269 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA383750071 rs1555148620 |
283 | D>N | No |
ClinGen gnomAD |
|
|
CA383750091 rs1371026118 |
284 | H>R | No |
ClinGen TOPMed |
|
|
rs1555148621 CA383750087 |
284 | H>Y | No |
ClinGen gnomAD |
|
|
rs1323488790 CA383750113 |
286 | R>Q | No |
ClinGen TOPMed |
|
|
CA232443973 rs56111095 |
287 | V>M | No |
ClinGen Ensembl |
|
|
CA383750166 rs1387683991 |
292 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs782600635 CA6421705 |
292 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs782502543 CA6421707 |
293 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1378668798 CA383750182 |
294 | S>G | No |
ClinGen TOPMed gnomAD |
|
|
rs782646307 CA6421708 |
296 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA6421712 rs782595592 |
298 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs782595592 CA6421711 |
298 | G>W | No |
ClinGen ExAC gnomAD |
|
|
rs754255009 CA232443997 |
300 | D>N | No |
ClinGen Ensembl |
|
|
rs1555148653 CA383750300 |
305 | N>S | No |
ClinGen gnomAD |
|
|
CA383750319 rs1252389772 |
307 | I>V | No |
ClinGen TOPMed |
|
|
rs1555148694 CA383750389 |
310 | Q>K | No |
ClinGen gnomAD |
|
|
rs1555148695 CA383750396 |
310 | Q>R | No |
ClinGen gnomAD |
|
|
rs1555148696 CA383750425 |
313 | G>D | No |
ClinGen Ensembl |
|
|
CA383750433 rs1555148698 |
314 | P>A | No |
ClinGen gnomAD |
|
|
CA383750434 rs1555148698 |
314 | P>S | No |
ClinGen gnomAD |
|
|
CA383750456 rs1198645982 |
316 | E>K | No |
ClinGen TOPMed |
|
|
CA6421736 rs782211008 |
318 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA383750485 rs1555148704 |
318 | A>V | No |
ClinGen gnomAD |
|
|
CA383750500 rs1591689003 |
320 | T>P | No |
ClinGen Ensembl |
|
|
rs782127411 CA6421739 |
322 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs1555148710 CA383750532 |
323 | A>T | No |
ClinGen gnomAD |
|
|
rs781923789 CA6421741 |
325 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA6421746 rs199933225 |
332 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs782547213 CA6421748 |
339 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201976465 CA6421749 |
339 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782608469 CA383750738 |
341 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
rs782608469 CA6421752 |
341 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA383750746 rs1591689121 |
342 | E>G | No |
ClinGen Ensembl |
|
|
rs369337036 CA6421753 |
345 | R>C | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
|
rs1470957233 CA383750773 |
345 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs879984166 CA383750780 |
346 | V>I | No |
ClinGen Ensembl |
|
|
rs782671210 CA6421755 |
348 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA383750801 rs1177433416 |
348 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA6421756 rs782187261 |
350 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs923201462 CA232444147 |
352 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1555148751 CA383750905 |
358 | R>Q | No |
ClinGen gnomAD |
|
|
CA383751190 rs1555148933 |
359 | N>K | No |
ClinGen gnomAD |
|
|
rs1555148930 CA383751186 |
359 | N>S | No |
ClinGen gnomAD |
|
|
CA6421786 rs781941676 |
360 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA383751199 rs1555148935 |
361 | C>S | No |
ClinGen gnomAD |
|
|
CA6421788 rs782761798 |
362 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1177506427 CA383751212 |
363 | P>T | No |
ClinGen TOPMed |
|
|
CA383751228 rs1555148944 |
365 | W>R | No |
ClinGen gnomAD |
|
|
CA232444495 rs959921236 |
367 | E>A | No |
ClinGen Ensembl |
|
|
COSM1512879 CA6421790 rs377517197 |
367 | E>K | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs377517197 CA6421791 |
367 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781806998 CA6421792 |
368 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA6421795 rs370538000 |
372 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370538000 CA6421794 |
372 | R>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782528840 CA6421796 |
372 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782674280 CA6421797 |
377 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA383751333 rs1484282642 |
381 | N>S | No |
ClinGen TOPMed |
|
|
rs782455819 CA6421799 |
383 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555148958 CA383751355 |
384 | E>D | No |
ClinGen gnomAD |
|
|
rs782570503 CA6421800 |
385 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA383751368 rs1555148961 |
386 | D>G | No |
ClinGen gnomAD |
|
|
rs376886181 CA6421803 |
389 | E>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782346134 CA6421802 |
389 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421804 rs782262226 |
390 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs782405424 CA6421805 |
392 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421806 COSM549666 rs782031352 |
393 | R>C | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs782044132 CA6421807 |
393 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1555148971 CA383751428 |
396 | Q>E | No |
ClinGen gnomAD |
|
|
rs1555148971 CA383751427 |
396 | Q>K | No |
ClinGen gnomAD |
|
|
CA6421808 rs782746802 |
398 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA6421809 rs781971151 |
399 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1302893456 CA383751461 |
401 | D>G | No |
ClinGen TOPMed |
|
|
CA383751501 rs1249604029 |
405 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
CA383751500 rs1249604029 |
405 | L>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs782681772 CA6421844 |
405 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs781934308 CA232444621 |
406 | I>M | No |
ClinGen Ensembl |
|
|
CA383751511 rs1489313099 |
407 | R>Q | No |
ClinGen TOPMed |
|
|
rs1555149024 CA383751623 |
422 | V>A | No |
ClinGen gnomAD |
|
|
rs781949874 CA6421848 |
424 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1555149029 CA383751658 |
427 | G>A | No |
ClinGen gnomAD |
|
|
CA6421850 rs782368432 |
428 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA6421851 rs781994700 |
428 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782142224 CA6421852 |
429 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1356734316 CA383751708 |
435 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1555149035 CA383751725 |
437 | N>K | No |
ClinGen gnomAD |
|
|
CA6421854 rs374365975 |
438 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA383751731 rs1555149037 |
438 | Q>R | No |
ClinGen gnomAD |
|
|
CA6421856 rs782749360 |
439 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421855 rs782065208 |
439 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs200005925 CA6421858 |
441 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781860095 CA6421857 |
441 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1305426435 CA383751751 |
442 | S>C | No |
ClinGen TOPMed |
|
|
rs1591690821 CA383751758 |
442 | S>R | No |
ClinGen Ensembl |
|
|
CA383751762 rs1565583883 |
443 | L>P | No |
ClinGen Ensembl |
|
|
CA6421859 rs368398907 |
443 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6421861 rs782437067 |
444 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs782216134 CA383751776 |
446 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6421864 rs782216134 |
446 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782216134 CA6421863 |
446 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782625225 CA6421865 |
446 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA6421867 rs782404245 |
448 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs782028376 CA6421868 |
449 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA383751797 rs1555149058 |
450 | I>V | No |
ClinGen gnomAD |
|
|
rs62621988 CA6421870 |
451 | V>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6421869 rs62621988 |
451 | V>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1363181410 CA383751826 |
454 | S>N | No |
ClinGen TOPMed |
|
|
rs1435682678 CA383751847 |
455 | A>S | No |
ClinGen TOPMed |
|
|
rs1435682678 CA383751849 |
455 | A>T | No |
ClinGen TOPMed |
|
|
rs1555149392 CA383751856 |
456 | G>D | No |
ClinGen gnomAD |
|
|
rs782000132 CA6421888 |
457 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1591692944 CA383751875 |
459 | R>P | No |
ClinGen Ensembl |
|
|
rs926689109 CA232447716 |
466 | I>S | No |
ClinGen Ensembl |
|
|
rs376994439 CA6421892 |
467 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1565584932 CA383751938 |
469 | L>V | No |
ClinGen Ensembl |
|
|
rs1591693004 CA383751976 |
474 | S>P | No |
ClinGen Ensembl |
|
|
CA6421893 rs782729581 |
475 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1171574303 CA383751990 |
476 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA383752097 rs1555149453 |
489 | Q>L | No |
ClinGen gnomAD |
|
|
rs1555149453 CA383752096 |
489 | Q>R | No |
ClinGen gnomAD |
|
|
rs1272459641 COSM943098 CA383752124 |
493 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1356852472 CA383752137 |
495 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA383752145 rs1283407292 |
496 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA232447860 rs782414151 |
501 | T>M | No |
ClinGen Ensembl |
|
|
CA6421932 rs185835888 |
503 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA383752216 rs1555149474 |
506 | K>N | No |
ClinGen gnomAD |
|
|
CA6421933 rs782252503 |
507 | F>L | No |
ClinGen ExAC |
|
|
CA6421936 rs782170877 |
510 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1471321885 CA383752272 |
515 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1555149485 CA383752283 |
516 | I>T | No |
ClinGen gnomAD |
|
|
CA6421938 rs781952099 |
519 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA383752342 rs1555149492 |
525 | V>I | No |
ClinGen gnomAD |
|
|
CA383752348 rs1555149495 |
526 | L>M | No |
ClinGen gnomAD |
|
|
COSM1263370 rs200635165 CA6422009 |
528 | S>L | oesophagus [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs200635165 CA383752378 |
528 | S>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555149599 CA383752381 |
529 | Q>* | No |
ClinGen gnomAD |
|
|
CA383752394 rs1555149602 |
530 | K>N | No |
ClinGen gnomAD |
|
|
CA383752425 rs1591693786 |
534 | S>A | No |
ClinGen Ensembl |
|
|
rs782688576 CA6422011 |
534 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1591693786 CA383752429 |
534 | S>P | No |
ClinGen Ensembl |
|
|
CA383752458 rs1555149614 |
536 | Y>C | No |
ClinGen gnomAD |
|
|
rs1009148404 CA232448104 |
536 | Y>H | No |
ClinGen gnomAD |
|
|
CA6422016 rs782494259 |
540 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA383752505 rs782494259 |
540 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA383752526 rs1370018915 |
542 | P>S | No |
ClinGen TOPMed |
|
|
CA383752539 rs1307380937 |
543 | P>Q | No |
ClinGen TOPMed |
|
|
rs1555149629 CA383752547 |
544 | A>P | No |
ClinGen gnomAD |
|
|
rs782265270 CA6422018 |
545 | M>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782265270 CA383752561 |
545 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383752558 rs1555149633 |
545 | M>V | No |
ClinGen gnomAD |
|
|
rs782487049 CA232448119 |
546 | K>R | No |
ClinGen gnomAD |
|
|
rs1555149641 CA383752606 |
548 | A>V | No |
ClinGen gnomAD |
|
|
CA6422019 rs782555031 |
551 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA6422020 rs782253557 |
552 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs782253557 CA383752645 |
552 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA232448127 rs782253557 |
552 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA6422021 rs200155138 |
553 | S>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM179640 rs782599498 CA6422023 |
554 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA6422024 rs782226517 |
554 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383752669 rs782226517 |
554 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1591693916 CA383752680 |
555 | T>I | No |
ClinGen Ensembl |
|
|
CA6422025 rs782366247 |
556 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555149653 CA383752709 |
558 | K>R | No |
ClinGen gnomAD |
|
|
CA383752708 rs1555149653 |
558 | K>T | No |
ClinGen gnomAD |
|
|
CA383753277 rs1555149806 |
559 | H>R | No |
ClinGen gnomAD |
|
|
rs1270713882 CA383753300 |
560 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
rs781928397 CA6422068 |
560 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1555149810 CA383753314 |
561 | E>D | No |
ClinGen gnomAD |
|
|
CA383753302 rs1591694546 |
561 | E>K | No |
ClinGen Ensembl |
|
|
CA383753364 rs1555149814 |
564 | Y>* | No |
ClinGen gnomAD |
|
|
rs782332287 CA6422070 |
567 | L>M | No |
ClinGen ExAC |
|
|
CA6422072 rs782106910 |
568 | H>P | No |
ClinGen ExAC |
|
|
rs1555149821 CA383753405 |
568 | H>Q | No |
ClinGen gnomAD |
|
|
rs375291511 CA6422071 |
568 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1231052417 CA383753410 |
569 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA232448409 rs782460041 |
571 | N>D | No |
ClinGen Ensembl |
|
|
CA232448412 rs978237552 |
572 | K>R | No |
ClinGen TOPMed |
|
|
CA383753437 rs978237552 |
572 | K>T | No |
ClinGen TOPMed |
|
|
rs1555149837 CA383753443 |
573 | R>G | No |
ClinGen gnomAD |
|
|
CA383753458 rs1555149838 |
574 | E>D | No |
ClinGen gnomAD |
|
|
rs1288467058 CA383753469 |
576 | K>E | No |
ClinGen TOPMed |
|
|
CA383753477 rs1555149841 |
577 | V>M | No |
ClinGen gnomAD |
|
|
CA6422073 rs782777814 |
580 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6422075 rs782167753 |
581 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782020090 CA6422074 |
581 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1326097283 CA383753514 |
582 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA6422079 rs367563603 |
585 | K>E | No |
ClinGen ESP ExAC TOPMed |
|
|
rs1555149852 CA383753534 |
585 | K>R | No |
ClinGen gnomAD |
|
|
CA383753558 rs1390422023 |
587 | K>E | No |
ClinGen TOPMed |
|
|
rs1163107482 CA383753567 |
587 | K>N | No |
ClinGen TOPMed |
|
|
CA383753573 rs1555149856 |
588 | S>G | No |
ClinGen gnomAD |
|
|
CA383753577 rs1555149857 |
588 | S>N | No |
ClinGen gnomAD |
|
|
CA6422082 rs781866962 |
590 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383753629 rs1555149861 |
591 | S>F | No |
ClinGen gnomAD |
|
|
CA6422083 rs782532946 |
592 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs782676287 CA383753652 |
593 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1468613421 CA383753663 |
594 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1555149866 CA383753668 |
595 | K>E | No |
ClinGen gnomAD |
No associated diseases with P29350
6 regional properties for P29350
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Tyrosine-specific protein phosphatase, PTPase domain | 243 - 517 | IPR000242 |
| domain | Tyrosine-specific protein phosphatases domain | 430 - 506 | IPR000387 |
| domain | SH2 domain | 2 - 100 | IPR000980-1 |
| domain | SH2 domain | 108 - 213 | IPR000980-2 |
| domain | Protein-tyrosine phosphatase, catalytic | 408 - 514 | IPR003595 |
| active_site | Protein-tyrosine phosphatase, active site | 451 - 461 | IPR016130 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.3.48 | Phosphoric monoester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR46257 | TYROSINE-PROTEIN PHOSPHATASE CORKSCREW |
| PANTHER Subfamily | PTHR46257:SF4 | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 6 |
| PANTHER Protein Class |
protein phosphatase
protein modifying enzyme |
|
| PANTHER Pathway Category |
B cell activation SHP-1 FGF signaling pathway SHP2 Angiogenesis SHP2 Interferon-gamma signaling pathway PTP |
|
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| alpha-beta T cell receptor complex | A T cell receptor complex in which the TCR heterodimer comprises alpha and beta chains, associated with the CD3 complex; recognizes a complex consisting of an antigen-derived peptide bound to a class I or class II MHC protein. |
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| specific granule lumen | The volume enclosed by the membrane of a specific granule, a granule with a membranous, tubular internal structure, found primarily in mature neutrophil cells. Most are released into the extracellular fluid. Specific granules contain lactoferrin, lysozyme, vitamin B12 binding protein and elastase. |
| tertiary granule lumen | Any membrane-enclosed lumen that is part of a tertiary granule. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| cell adhesion molecule binding | Binding to a cell adhesion molecule. |
| phosphorylation-dependent protein binding | Binding to a protein upon phosphorylation of the target protein. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein tyrosine phosphatase activity | Catalysis of the reaction: protein tyrosine phosphate + H2O = protein tyrosine + phosphate. |
| SH2 domain binding | Binding to a SH2 domain (Src homology 2) of a protein, a protein domain of about 100 amino-acid residues and belonging to the alpha + beta domain class. |
| SH3 domain binding | Binding to a SH3 domain (Src homology 3) of a protein, small protein modules containing approximately 50 amino acid residues found in a great variety of intracellular or membrane-associated proteins. |
| transmembrane receptor protein tyrosine phosphatase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: protein tyrosine phosphate + H2O = protein tyrosine + phosphate. |
34 GO annotations of biological process
| Name | Definition |
|---|---|
| abortive mitotic cell cycle | A cell cycle in which mitosis is begun and progresses normally through the end of anaphase, but not completed, resulting in a cell with increased ploidy. |
| B cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a B cell. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cytokine-mediated signaling pathway | The series of molecular signals initiated by the binding of a cytokine to a receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| epididymis development | The process whose specific outcome is the progression of an epididymis over time, from its formation to the mature structure. |
| G protein-coupled receptor signaling pathway | The series of molecular signals initiated by a ligand binding to its receptor, in which the activated receptor promotes the exchange of GDP for GTP on the alpha-subunit of an associated heterotrimeric G-protein complex. The GTP-bound activated alpha-G-protein then dissociates from the beta- and gamma-subunits to further transmit the signal within the cell. The pathway begins with receptor-ligand interaction, and ends with regulation of a downstream cellular process. The pathway can start from the plasma membrane, Golgi or nuclear membrane. |
| hematopoietic progenitor cell differentiation | The process in which precursor cell type acquires the specialized features of a hematopoietic progenitor cell, a class of cell types including myeloid progenitor cells and lymphoid progenitor cells. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| megakaryocyte development | The process whose specific outcome is the progression of a megakaryocyte cell over time, from its formation to the mature structure. Megakaryocyte development does not include the steps involved in committing a cell to a megakaryocyte fate. A megakaryocyte is a giant cell 50 to 100 micron in diameter, with a greatly lobulated nucleus, found in the bone marrow. |
| natural killer cell mediated cytotoxicity | The directed killing of a target cell by a natural killer cell through the release of granules containing cytotoxic mediators or through the engagement of death receptors. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of humoral immune response mediated by circulating immunoglobulin | Any process that stops, prevents, or reduces the frequency, rate, or extent of a humoral immune response mediated by circulating immunoglobulin. |
| negative regulation of interleukin-6 production | Any process that stops, prevents, or reduces the frequency, rate, or extent of interleukin-6 production. |
| negative regulation of MAP kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of MAP kinase activity. |
| negative regulation of mast cell activation involved in immune response | Any process that stops, prevents, or reduces the frequency, rate, or extent of mast cell activation as part of an immune response. |
| negative regulation of peptidyl-tyrosine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
| negative regulation of T cell proliferation | Any process that stops, prevents or reduces the rate or extent of T cell proliferation. |
| negative regulation of T cell receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of signaling pathways initiated by the cross-linking of an antigen receptor on a T cell. |
| negative regulation of tumor necrosis factor production | Any process that stops, prevents, or reduces the frequency, rate, or extent of tumor necrosis factor production. |
| peptidyl-tyrosine dephosphorylation | The removal of phosphoric residues from peptidyl-O-phospho-tyrosine to form peptidyl-tyrosine. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| platelet aggregation | The adhesion of one platelet to one or more other platelets via adhesion molecules. |
| platelet formation | The process in which platelets bud from long processes extended by megakaryocytes. |
| positive regulation of cell adhesion mediated by integrin | Any process that activates or increases the frequency, rate, or extent of cell adhesion mediated by integrin. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of phosphatidylinositol 3-kinase signaling | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the phosphatidylinositol 3-kinase cascade. |
| protein dephosphorylation | The process of removing one or more phosphoric residues from a protein. |
| regulation of apoptotic process | Any process that modulates the occurrence or rate of cell death by apoptotic process. |
| regulation of B cell differentiation | Any process that modulates the frequency, rate or extent of B cell differentiation. |
| regulation of ERK1 and ERK2 cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| regulation of G1/S transition of mitotic cell cycle | Any signalling pathway that modulates the activity of a cell cycle cyclin-dependent protein kinase to modulate the switch from G1 phase to S phase of the mitotic cell cycle. |
| regulation of release of sequestered calcium ion into cytosol | Any process that modulates the frequency, rate or extent of the release into the cytosolic compartment of calcium ions sequestered in the endoplasmic reticulum or mitochondria. |
| regulation of type I interferon-mediated signaling pathway | Any process that modulates the rate, frequency or extent of a type I interferon-mediated signaling pathway. |
| T cell costimulation | The process of providing, via surface-bound receptor-ligand pairs, a second, antigen-independent, signal in addition to that provided by the T cell receptor to augment T cell activation. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q90687 | PTPN11 | Tyrosine-protein phosphatase non-receptor type 11 | Gallus gallus (Chicken) | SS |
| Q06124 | PTPN11 | Tyrosine-protein phosphatase non-receptor type 11 | Homo sapiens (Human) | EV |
| P35235 | Ptpn11 | Tyrosine-protein phosphatase non-receptor type 11 | Mus musculus (Mouse) | SS |
| P29351 | Ptpn6 | Tyrosine-protein phosphatase non-receptor type 6 | Mus musculus (Mouse) | SS |
| P41499 | Ptpn11 | Tyrosine-protein phosphatase non-receptor type 11 | Rattus norvegicus (Rat) | SS |
| P81718 | Ptpn6 | Tyrosine-protein phosphatase non-receptor type 6 | Rattus norvegicus (Rat) | SS |
| G5EC24 | ptp-2 | Tyrosine-protein phosphatase non-receptor type ptp-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVRWFHRDLS | GLDAETLLKG | RGVHGSFLAR | PSRKNQGDFS | LSVRVGDQVT | HIRIQNSGDF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YDLYGGEKFA | TLTELVEYYT | QQQGVLQDRD | GTIIHLKYPL | NCSDPTSERW | YHGHMSGGQA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ETLLQAKGEP | WTFLVRESLS | QPGDFVLSVL | SDQPKAGPGS | PLRVTHIKVM | CEGGRYTVGG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LETFDSLTDL | VEHFKKTGIE | EASGAFVYLR | QPYYATRVNA | ADIENRVLEL | NKKQESEDTA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KAGFWEEFES | LQKQEVKNLH | QRLEGQRPEN | KGKNRYKNIL | PFDHSRVILQ | GRDSNIPGSD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YINANYIKNQ | LLGPDENAKT | YIASQGCLEA | TVNDFWQMAW | QENSRVIVMT | TREVEKGRNK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| CVPYWPEVGM | QRAYGPYSVT | NCGEHDTTEY | KLRTLQVSPL | DNGDLIREIW | HYQYLSWPDH |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GVPSEPGGVL | SFLDQINQRQ | ESLPHAGPII | VHCSAGIGRT | GTIIVIDMLM | ENISTKGLDC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DIDIQKTIQM | VRAQRSGMVQ | TEAQYKFIYV | AIAQFIETTK | KKLEVLQSQK | GQESEYGNIT |
| 550 | 560 | 570 | 580 | 590 | |
| YPPAMKNAHA | KASRTSSKHK | EDVYENLHTK | NKREEKVKKQ | RSADKEKSKG | SLKRK |