P29322
Gene name |
EPHA8 (EEK, HEK3, KIAA1459) |
Protein name |
Ephrin type-A receptor 8 |
Names |
EC 2.7.10.1 , EPH- and ELK-related kinase , EPH-like kinase 3 , EK3 , hEK3 , Tyrosine-protein kinase receptor EEK |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2046 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
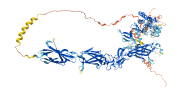
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
635-896 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
777-803 (Activation loop from InterPro)
Target domain |
635-896 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
Autoinhibited structure

Activated structure

4 structures for P29322
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1UCV | NMR | - | A | 933-1000 | PDB |
| 1X5L | NMR | - | A | 437-534 | PDB |
| 3KUL | X-ray | 215 A | A/B | 602-909 | PDB |
| AF-P29322-F1 | Predicted | AlphaFoldDB |
1269 variants for P29322
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001572900 RCV002569063 rs150553424 |
93 | G>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1640264975 | 2 | A>V | No | TOPMed | |
| rs1640265044 | 3 | P>A | No | Ensembl | |
| rs1640265107 | 4 | A>T | No | Ensembl | |
| rs1166883643 | 5 | R>W | No |
TOPMed gnomAD |
|
| rs2124501802 | 6 | G>A | No | Ensembl | |
| rs1042555143 | 7 | R>C | No | gnomAD | |
| rs902613029 | 7 | R>H | No |
TOPMed gnomAD |
|
| rs902613029 | 7 | R>P | No |
TOPMed gnomAD |
|
| rs1309912365 | 9 | P>L | No | Ensembl | |
| rs1309912365 | 9 | P>R | No | Ensembl | |
| rs1640266007 | 9 | P>S | No | gnomAD | |
| rs1640266268 | 10 | P>A | No | gnomAD | |
| rs1640266390 | 10 | P>R | No | TOPMed | |
| rs1640266268 | 10 | P>T | No | gnomAD | |
| rs1311276052 | 11 | A>E | No | gnomAD | |
| rs1209020245 | 11 | A>P | No | TOPMed | |
| rs1209020245 | 11 | A>T | No | TOPMed | |
| rs1640267152 | 13 | W>* | No | TOPMed | |
| rs1640267415 | 16 | T>K | No | TOPMed | |
| rs1640268110 | 19 | A>V | No | Ensembl | |
| rs2124501904 | 20 | A>G | No | Ensembl | |
| rs2124501912 | 21 | A>G | No | Ensembl | |
| rs1640268336 | 21 | A>S | No | 1000Genomes | |
| rs1640268396 | 23 | T>P | No | TOPMed | |
| rs1234772137 | 25 | V>L | No | gnomAD | |
| rs1483495299 | 28 | A>E | No | TOPMed | |
| rs1483495299 | 28 | A>V | No | TOPMed | |
| rs1640268864 | 29 | R>P | No | Ensembl | |
| TCGA novel | 30 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557546661 | 30 | G>S | No | Ensembl | |
| rs765990373 | 34 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs773672647 | 35 | L>V | No |
ExAC TOPMed gnomAD |
|
|
COSM464094 COSM464093 |
36 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200476235 | 37 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs200476235 | 37 | T>M | No |
ExAC TOPMed gnomAD |
|
|
COSM464098 COSM464097 |
37 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767316353 | 38 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1052037972 | 38 | S>P | No | TOPMed | |
| rs767316353 | 38 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs1383954348 | 39 | T>A | No |
TOPMed gnomAD |
|
| rs1383954348 | 39 | T>P | No |
TOPMed gnomAD |
|
| rs147107186 | 40 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs763862668 | 41 | H>Y | No |
ExAC gnomAD |
|
| rs1329533785 | 42 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs370205666 | 45 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
|
VAR_042153 rs45498698 |
45 | G>S | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs370205666 | 45 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM266468 COSM266467 |
46 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1242022682 | 47 | L>P | No | gnomAD | |
| rs367682437 | 48 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs367682437 | 48 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs367682437 | 48 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs777065034 | 50 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1480515620 | 51 | A>S | No |
TOPMed gnomAD |
|
|
rs1640457971 TCGA novel |
53 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
COSM3484869 COSM3484868 |
54 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745520846 | 54 | W>G | No |
ExAC TOPMed gnomAD |
|
| rs1419113927 | 54 | W>L | No | gnomAD | |
| rs1290026909 | 55 | D>A | No |
TOPMed gnomAD |
|
| rs1429380962 | 55 | D>E | No | gnomAD | |
| rs1640686975 | 55 | D>N | No | TOPMed | |
| rs1290026909 | 55 | D>V | No |
TOPMed gnomAD |
|
| rs2124519825 | 57 | I>M | No | Ensembl | |
| rs1172776048 | 57 | I>S | No | gnomAD | |
| rs771547728 | 58 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs771547728 | 58 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs925019483 | 59 | E>A | No | TOPMed | |
| rs768533785 | 59 | E>D | No |
ExAC gnomAD |
|
| rs760621090 | 59 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
rs56402644 VAR_042154 |
60 | V>L | No |
UniProt Ensembl dbSNP |
|
| rs1557555588 | 61 | D>N | No | Ensembl | |
| rs142067038 | 62 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
|
rs142067038 COSM3484871 COSM3484870 |
62 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs750672863 COSM3864712 COSM3864713 |
63 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1569962417 | 63 | S>P | No | Ensembl | |
| rs766378509 | 65 | Q>* | No |
ExAC gnomAD |
|
| rs1233938022 | 65 | Q>R | No | gnomAD | |
| rs751639041 | 66 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1640689096 | 66 | P>R | No | gnomAD | |
| rs751639041 | 66 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs2124519914 | 68 | H>R | No | Ensembl | |
| rs755571143 | 68 | H>Y | No |
ExAC gnomAD |
|
|
rs1359234020 COSM305914 COSM305915 |
69 | T>M | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs753231159 | 73 | C>S | No |
ExAC gnomAD |
|
| rs2124519930 | 74 | N>S | No | Ensembl | |
| rs778195570 | 75 | V>I | No |
ExAC gnomAD |
|
| rs1484910204 | 76 | M>I | No |
TOPMed gnomAD |
|
| rs1204795436 | 77 | S>R | No | TOPMed | |
| rs577503096 | 77 | S>R | No |
1000Genomes ExAC gnomAD |
|
| rs1257051181 | 78 | P>A | No | TOPMed | |
| rs771697649 | 78 | P>L | No |
ExAC gnomAD |
|
| rs1640690627 | 79 | N>D | No | Ensembl | |
|
COSM1687168 rs372331897 COSM1687169 |
79 | N>S | skin [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1436544769 | 80 | Q>E | No | gnomAD | |
| rs376900516 | 80 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1344147370 | 81 | N>H | No |
TOPMed gnomAD |
|
| rs544912245 | 81 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1344147370 | 81 | N>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 84 | L>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs567786868 COSM905204 |
85 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs763102049 | 85 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs763102049 | 85 | R>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 86 | T>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200156030 | 86 | T>M | No |
1000Genomes ExAC gnomAD |
|
| rs759737535 | 87 | S>N | No |
ExAC gnomAD |
|
| rs1402471074 | 88 | W>* | No |
TOPMed gnomAD |
|
| rs1402471074 | 88 | W>C | No |
TOPMed gnomAD |
|
| rs767689378 | 90 | P>S | No |
ExAC gnomAD |
|
| rs767689378 | 90 | P>T | No |
ExAC gnomAD |
|
| rs753322537 | 91 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs753322537 | 91 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs756576145 | 91 | R>P | No |
ExAC gnomAD |
|
|
COSM1997277 COSM1997278 rs756576145 |
91 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs145476843 | 92 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150553424 | 93 | G>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1455408456 | 93 | G>D | No | gnomAD | |
| rs1455408456 | 93 | G>V | No | gnomAD | |
|
COSM2154345 COSM2154346 rs758018689 |
94 | A>T | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs148846897 | 95 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM464100 rs751977374 COSM464099 |
95 | R>W | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1391875727 COSM905206 |
96 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1391875727 | 96 | R>G | No | gnomAD | |
| rs754544338 | 96 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs562198376 COSM42640 COSM1559280 |
97 | V>I | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs748046093 | 98 | Y>H | No |
ExAC gnomAD |
|
| rs951196362 | 99 | A>G | No | TOPMed | |
| rs769718758 | 100 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs1374620571 | 100 | E>D | No |
TOPMed gnomAD |
|
|
COSM3721646 rs769718758 COSM3721645 |
100 | E>K | upper_aerodigestive_tract Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC TOPMed gnomAD |
| rs1177727145 | 101 | I>T | No | Ensembl | |
| rs148164803 | 102 | K>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148164803 | 102 | K>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749144133 | 102 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs749144133 | 102 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1557555997 | 105 | L>Q | No | TOPMed | |
| TCGA novel | 105 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1339948 COSM1339949 |
106 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770825298 | 106 | R>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3484872 rs774769663 COSM3484873 |
107 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs774769663 | 107 | D>Y | No |
ExAC gnomAD |
|
| rs1470735573 | 109 | N>S | No |
TOPMed gnomAD |
|
| rs1232257889 | 111 | M>I | No |
TOPMed gnomAD |
|
| rs1179328822 | 111 | M>V | No | gnomAD | |
| rs1569963170 | 112 | P>L | No | Ensembl | |
| rs759833113 | 113 | G>D | No |
ExAC gnomAD |
|
| rs1640696690 | 113 | G>R | No |
TOPMed gnomAD |
|
| rs767709429 | 114 | V>A | No |
ExAC gnomAD |
|
| rs1640697111 | 115 | L>P | No | gnomAD | |
| rs150187218 | 115 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1640697178 | 116 | G>S | No | Ensembl | |
| rs1165199164 | 116 | G>V | No | gnomAD | |
| rs764624482 | 117 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1408592467 | 117 | T>I | No |
TOPMed gnomAD |
|
| rs1408592467 | 117 | T>N | No |
TOPMed gnomAD |
|
| rs1302045042 | 118 | C>Y | No | TOPMed | |
| rs1569963294 | 122 | F>C | No | Ensembl | |
| VAR_042155 | 123 | N>K | a breast infiltrating ductal carcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs757692138 | 125 | Y>C | No |
ExAC gnomAD |
|
| rs905212002 | 126 | Y>C | No | TOPMed | |
| rs1640698322 | 126 | Y>D | No | Ensembl | |
| rs369145872 | 128 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1019479070 | 129 | S>A | No | Ensembl | |
| rs751283643 | 129 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs754562491 | 130 | D>V | No |
ExAC gnomAD |
|
|
rs757672919 COSM3984809 COSM3984810 |
131 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2124520469 | 131 | R>S | No | Ensembl | |
| TCGA novel | 132 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780699593 | 132 | D>N | No |
ExAC gnomAD |
|
| rs138738837 | 133 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3484879 rs1479607175 COSM3484878 |
134 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs146324097 | 134 | G>R | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 135 | A>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777812539 | 135 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM94206 rs777812539 |
135 | A>T | breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM3360728 COSM3360729 |
135 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs975474573 | 138 | Q>P | No | Ensembl | |
|
rs1474243277 COSM1997287 COSM1997286 |
139 | E>Q | urinary_tract [Cosmic] | No |
cosmic curated gnomAD |
| rs1166941280 | 141 | Q>R | No | gnomAD | |
| rs1333715861 | 143 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs774222703 | 143 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1411998753 | 144 | K>R | No |
TOPMed gnomAD |
|
| rs746101749 | 145 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs775464367 | 146 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs775464367 | 146 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs768600015 | 148 | I>T | No |
ExAC gnomAD |
|
| rs760851429 | 148 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs777255535 | 149 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs777255535 | 149 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1025510947 | 150 | A>T | No | TOPMed | |
| rs750733199 | 151 | D>E | No |
ExAC gnomAD |
|
|
COSM905209 COSM905210 |
151 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1278129338 | 151 | D>N | No |
TOPMed gnomAD |
|
| rs1278129338 | 151 | D>Y | No |
TOPMed gnomAD |
|
|
COSM5966986 rs1640702577 COSM5966985 |
152 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs1247192999 | 153 | S>N | No | gnomAD | |
| rs199987810 | 154 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199987810 | 154 | F>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767194142 | 155 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1640702971 | 155 | T>R | No | Ensembl | |
| rs752266814 | 156 | G>D | No |
ExAC gnomAD |
|
| rs1214998987 | 157 | A>T | No |
TOPMed gnomAD |
|
| rs1640703306 | 158 | D>N | No | TOPMed | |
| rs1640703969 | 159 | L>V | No | gnomAD | |
| rs370843084 | 160 | G>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370843084 | 160 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1451737171 | 161 | V>L | No | gnomAD | |
| rs1395045322 | 162 | R>P | No |
TOPMed gnomAD |
|
| rs1395045322 | 162 | R>Q | No |
TOPMed gnomAD |
|
| rs796756543 | 162 | R>W | No |
TOPMed gnomAD |
|
|
COSM414581 COSM414580 rs757202575 |
163 | R>H | urinary_tract [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs757202575 | 163 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1382969559 | 165 | K>E | No |
TOPMed gnomAD |
|
| rs142239276 | 168 | T>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs142239276 | 168 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs142239276 | 168 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1557556550 | 170 | V>M | No | Ensembl | |
|
COSM183246 COSM183245 rs768851018 |
171 | R>C | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs768851018 | 171 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs776511588 COSM905211 COSM905212 |
171 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs768851018 | 171 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1241219559 | 172 | S>G | No |
TOPMed gnomAD |
|
| rs1241219559 | 172 | S>R | No |
TOPMed gnomAD |
|
| rs570237058 | 172 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1640706090 | 174 | G>S | No | Ensembl | |
| rs773673708 | 175 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs773673708 | 175 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1257688669 | 175 | P>S | No | gnomAD | |
| rs766636647 | 176 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1188000635 | 177 | S>C | No | gnomAD | |
|
rs1557556639 COSM1639746 COSM20619 VAR_042156 |
179 | R>C | urinary_tract stomach a gastric adenocarcinoma sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated UniProt TOPMed dbSNP gnomAD |
| rs760177071 | 179 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs187814118 | 180 | G>C | No |
1000Genomes ESP ExAC gnomAD |
|
| rs187814118 | 180 | G>S | No |
1000Genomes ESP ExAC gnomAD |
|
| rs145031815 | 181 | F>Y | No |
ESP TOPMed |
|
| rs1569964107 | 182 | Y>S | No | Ensembl | |
| rs1370039670 | 183 | L>V | No | gnomAD | |
| rs753237688 | 184 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1227307074 | 186 | Q>R | No |
TOPMed gnomAD |
|
| rs1453171448 | 187 | D>N | No | gnomAD | |
| rs1387189639 | 188 | I>M | No | gnomAD | |
| rs1640709348 | 188 | I>R | No |
TOPMed gnomAD |
|
| rs1640709348 | 188 | I>T | No |
TOPMed gnomAD |
|
| rs779034408 | 188 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1361482584 | 189 | G>R | No | TOPMed | |
| rs768686298 | 190 | A>S | No |
ExAC gnomAD |
|
| rs768686298 | 190 | A>T | No |
ExAC gnomAD |
|
| rs1325451914 | 191 | C>R | No | gnomAD | |
| rs1400353622 | 192 | L>M | No |
TOPMed gnomAD |
|
| rs1640710151 | 192 | L>R | No | TOPMed | |
| rs1400353622 | 192 | L>V | No |
TOPMed gnomAD |
|
| rs758328561 | 194 | I>V | No | ExAC | |
|
COSM4896507 COSM4896506 rs1466759802 |
196 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1333861109 | 198 | R>C | No |
TOPMed gnomAD |
|
|
rs370576065 COSM905213 COSM905214 |
198 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| VAR_042157 | 198 | R>L | a lung adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
| TCGA novel | 198 | R>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1330578538 | 199 | I>M | No |
TOPMed gnomAD |
|
| rs1260845914 | 199 | I>V | No | gnomAD | |
| rs1640711322 | 201 | Y>* | No | Ensembl | |
| rs747263088 | 201 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs781359092 | 204 | C>R | No |
ExAC gnomAD |
|
| rs769840240 | 205 | P>H | No |
ExAC gnomAD |
|
| rs1640712160 | 206 | A>T | No | Ensembl | |
| rs749789776 | 207 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1640712587 | 209 | R>C | No | gnomAD | |
| rs62642541 | 209 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1640712821 | 210 | N>D | No | TOPMed | |
| rs1640712921 | 210 | N>S | No |
TOPMed gnomAD |
|
|
COSM1743270 rs56193119 COSM1743269 |
211 | L>M | biliary_tract [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs776392628 | 215 | S>L | No |
ExAC TOPMed gnomAD |
|
|
COSM1718751 COSM1718752 |
216 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759801337 | 217 | A>T | No |
ExAC gnomAD |
|
| rs764828706 | 218 | V>A | No |
ExAC gnomAD |
|
| TCGA novel | 219 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1394031937 COSM4029295 COSM4029294 |
219 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM3484882 COSM3484883 |
220 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1390798586 | 221 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1640713975 | 221 | A>V | No | TOPMed | |
|
COSM905215 rs1351802924 COSM905216 |
222 | D>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs758347205 | 223 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs779712228 | 224 | S>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 224 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs544874360 | 227 | V>M | No | 1000Genomes | |
| TCGA novel | 230 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1640715462 | 231 | G>S | No | Ensembl | |
| rs1223268633 | 232 | Q>R | No |
TOPMed gnomAD |
|
| rs1384956238 | 233 | C>F | No | TOPMed | |
| rs556852015 | 234 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1733943 COSM1733944 rs556852015 |
234 | V>M | pancreas [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs374015787 | 235 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs374015787 | 235 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs748354273 COSM1997309 COSM1997310 |
235 | R>W | ovary [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs372232524 | 236 | H>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs201777623 | 239 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs746259616 | 240 | R>Q | No |
ExAC gnomAD |
|
|
rs202149500 COSM5443170 COSM5443169 |
240 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1423915318 | 241 | D>A | No |
TOPMed gnomAD |
|
|
COSM905218 COSM905217 |
241 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4404882 COSM4404883 |
243 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1308574178 | 243 | P>S | No | gnomAD | |
| rs776485975 | 245 | M>I | No |
ExAC gnomAD |
|
| rs975314947 | 245 | M>R | No |
TOPMed gnomAD |
|
| rs761554734 | 248 | S>G | No |
ExAC TOPMed gnomAD |
|
|
COSM425540 rs202015824 COSM425541 |
249 | A>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
|
rs768374859 COSM2156533 COSM2156534 |
249 | A>V | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1173631731 | 250 | E>D | No | gnomAD | |
| rs1640719004 | 250 | E>G | No | TOPMed | |
| rs1569965022 | 250 | E>K | No | Ensembl | |
|
COSM4823159 COSM4823158 |
250 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766263755 | 251 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1640719498 | 253 | W>* | No | Ensembl | |
| rs368167619 | 254 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs368167619 | 254 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs368167619 | 254 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs752919461 | 255 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs752919461 | 255 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1210758652 | 256 | P>S | No |
TOPMed gnomAD |
|
| rs1210758652 | 256 | P>T | No |
TOPMed gnomAD |
|
| TCGA novel | 257 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756270156 | 258 | G>S | No |
ExAC gnomAD |
|
| rs1484269942 | 259 | K>I | No |
TOPMed gnomAD |
|
| rs1484269942 | 259 | K>R | No |
TOPMed gnomAD |
|
|
COSM6124676 COSM6124677 |
260 | C>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749326341 | 260 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs757329498 | 261 | V>A | No |
ExAC gnomAD |
|
| rs1640721034 | 261 | V>M | No | TOPMed | |
| TCGA novel | 262 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746375082 | 263 | S>G | No |
ExAC gnomAD |
|
| rs1172918635 | 263 | S>R | No | gnomAD | |
| rs772385874 | 265 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs772385874 | 265 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs138569134 | 267 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3484884 COSM3484885 |
268 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3984814 COSM3984813 |
269 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139777546 | 269 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs747819394 | 269 | R>W | No |
ExAC TOPMed gnomAD |
|
|
COSM3484887 rs201689882 COSM3484886 |
270 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA gnomAD |
| rs1640722621 | 271 | D>E | No | TOPMed | |
| rs76874545 | 271 | D>G | No | Ensembl | |
| rs762414738 | 273 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs762414738 | 273 | C>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3484889 COSM3484888 |
274 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770458620 | 274 | V>G | No |
ExAC gnomAD |
|
| rs754009354 | 275 | A>D | No |
ExAC gnomAD |
|
|
COSM4029297 COSM4029296 |
275 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1279480547 | 277 | E>D | No | gnomAD | |
| rs1440723185 | 277 | E>K | No | gnomAD | |
| rs762088600 | 279 | G>A | No |
ExAC gnomAD |
|
| rs1641211110 | 280 | F>L | No | TOPMed | |
| rs1641211200 | 281 | Y>C | No | TOPMed | |
| rs147152756 | 282 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs750491008 | 283 | S>A | No |
ExAC gnomAD |
|
|
COSM459554 COSM459555 |
283 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1641211553 | 284 | A>G | No | gnomAD | |
| rs759009996 | 285 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1405311325 | 287 | D>N | No |
TOPMed gnomAD |
|
| rs1405311325 | 287 | D>Y | No |
TOPMed gnomAD |
|
| rs368255315 | 288 | Q>R | No |
ESP TOPMed gnomAD |
|
|
COSM533327 COSM6061499 rs920922969 COSM6061500 COSM533328 |
289 | L>R | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs752059573 | 292 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs752059573 | 292 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs755402558 | 292 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs752059573 | 292 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1641212677 | 293 | C>R | No | TOPMed | |
| rs1641212762 | 293 | C>Y | No | TOPMed | |
| rs1641212841 | 294 | P>H | No | Ensembl | |
|
COSM3484892 COSM3484893 |
294 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs557217944 | 295 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs557217944 | 295 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748953848 | 296 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs1474704377 | 296 | H>P | No | gnomAD | |
| rs748953848 | 296 | H>Y | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 297 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs141028919 | 300 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs141028919 | 300 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs947558247 | 301 | A>T | No | Ensembl | |
| rs638524 | 301 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs903455856 | 302 | P>L | No |
TOPMed gnomAD |
|
| rs903455856 | 302 | P>R | No |
TOPMed gnomAD |
|
| rs746880859 | 304 | A>S | No |
ExAC TOPMed gnomAD |
|
|
rs746880859 COSM679210 COSM679211 |
304 | A>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1246802459 | 305 | Q>H | No | TOPMed | |
| rs1641214671 | 306 | A>G | No |
TOPMed gnomAD |
|
| rs1354882287 | 306 | A>P | No | gnomAD | |
| rs1245880814 | 311 | L>F | No | gnomAD | |
| rs768504842 | 312 | S>C | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 313 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776470892 | 315 | R>C | No |
ExAC gnomAD |
|
|
rs761987517 COSM1748207 COSM1748208 |
315 | R>H | urinary_tract [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1273858169 | 317 | A>T | No |
TOPMed gnomAD |
|
| rs1570001321 | 319 | D>A | No | Ensembl | |
|
COSM1559281 rs765465256 COSM1559282 |
320 | P>L | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| COSM679209 | 320 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1207181815 | 320 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs56656925 VAR_061292 |
321 | P>L | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1557569561 | 325 | C>S | No | Ensembl | |
| rs1641216854 | 326 | T>N | No | Ensembl | |
|
rs929755154 COSM6124674 COSM1501234 COSM1501235 COSM6124675 |
327 | R>W | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1268297144 | 330 | S>L | No | gnomAD | |
| rs1557571697 | 331 | A>T | No | Ensembl | |
| rs200917073 | 331 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3864715 rs1641291735 COSM3864714 |
332 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs117363297 | 334 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1374109535 | 336 | I>V | No | TOPMed | |
| rs201043307 | 337 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs201043307 | 337 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1641292392 | 338 | S>N | No | Ensembl | |
| rs781208596 | 338 | S>R | No |
ExAC gnomAD |
|
| rs1362201512 | 339 | V>M | No |
TOPMed gnomAD |
|
| rs2148252704 | 340 | N>S | No | Ensembl | |
| rs769701249 | 341 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs1323688249 | 342 | T>A | No |
TOPMed gnomAD |
|
| rs375912312 | 342 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs202187898 | 344 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774675715 | 345 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1641293298 | 345 | T>S | No | gnomAD | |
| rs1333485762 | 346 | L>Q | No |
TOPMed gnomAD |
|
| rs1227183854 | 347 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1227183854 | 347 | E>K | No |
TOPMed gnomAD |
|
| rs571199591 | 347 | E>V | No |
1000Genomes ExAC gnomAD |
|
| rs1641293774 | 348 | W>G | No | gnomAD | |
| rs1641293856 | 349 | A>P | No | gnomAD | |
|
COSM1251355 COSM1251354 rs184229539 |
349 | A>V | oesophagus [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1641294154 | 350 | P>R | No | TOPMed | |
| rs2148252794 | 350 | P>S | No | Ensembl | |
| rs1052416939 | 351 | P>T | No | Ensembl | |
| rs1238316284 | 352 | L>P | No | gnomAD | |
| rs1641294635 | 353 | D>N | No | gnomAD | |
| rs2148252827 | 353 | D>V | No | Ensembl | |
| rs1641294716 | 354 | P>S | No | Ensembl | |
| rs1009368131 | 355 | G>A | No | TOPMed | |
| rs761081783 | 356 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1248963963 | 357 | R>C | No |
TOPMed gnomAD |
|
| rs373378884 | 357 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM168754 COSM168753 rs373378884 |
357 | R>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1248963963 | 357 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1414232134 | 359 | D>N | No |
TOPMed gnomAD |
|
| rs1414232134 | 359 | D>Y | No |
TOPMed gnomAD |
|
| rs762631777 | 360 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1641295491 | 362 | Y>C | No | Ensembl | |
|
COSM6124673 COSM6124672 |
362 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766048193 | 363 | N>D | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 363 | N>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766048193 | 363 | N>Y | No |
ExAC TOPMed gnomAD |
|
| rs866837685 | 364 | A>V | No | Ensembl | |
| rs369589341 | 365 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1408724603 | 366 | C>R | No |
TOPMed gnomAD |
|
|
COSM1559283 COSM42641 rs373048699 |
367 | R>C | central_nervous_system [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs755985438 | 367 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs777753788 | 368 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1311483721 | 368 | R>H | No |
TOPMed gnomAD |
|
| rs1322711881 | 369 | C>* | No | gnomAD | |
| rs1641297624 | 371 | W>* | No | gnomAD | |
| rs749128937 | 371 | W>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 372 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1277844699 | 372 | A>G | No | Ensembl | |
| TCGA novel | 372 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1250156610 | 372 | A>T | No |
TOPMed gnomAD |
|
| rs770802540 | 373 | L>P | No |
ExAC gnomAD |
|
| rs779291992 | 374 | S>I | No |
ExAC gnomAD |
|
| rs752982112 | 375 | R>C | No |
TOPMed gnomAD |
|
| rs201818748 | 375 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs201818748 | 375 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1257057563 | 377 | E>* | No |
TOPMed gnomAD |
|
| rs1257057563 | 377 | E>K | No |
TOPMed gnomAD |
|
| rs918958008 | 378 | A>V | No | Ensembl | |
| rs1641298817 | 380 | G>R | No | TOPMed | |
| rs1388058227 | 382 | G>A | No |
TOPMed gnomAD |
|
| rs1388058227 | 382 | G>D | No |
TOPMed gnomAD |
|
|
rs769118581 COSM166548 COSM166547 |
382 | G>S | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs372845574 | 383 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1172302228 | 384 | R>C | No |
TOPMed gnomAD |
|
| rs762185124 | 384 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs571969130 | 388 | Q>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759101185 | 390 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs767106998 | 391 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1379388674 | 391 | S>T | No | gnomAD | |
| rs755612112 | 393 | V>G | No |
ExAC gnomAD |
|
| COSM679207 | 393 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1328150773 | 393 | V>M | No |
TOPMed gnomAD |
|
| rs1557572182 | 396 | S>I | No | Ensembl | |
|
COSM4401205 COSM4401204 |
399 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757122925 | 400 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1350698742 | 401 | N>K | No | gnomAD | |
| rs2148253193 | 402 | L>M | No | Ensembl | |
| rs751900514 | 404 | A>T | No |
TOPMed gnomAD |
|
| rs1459701561 | 404 | A>V | No | gnomAD | |
| rs1641301722 | 405 | H>Y | No | TOPMed | |
| rs377088112 | 408 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553147565 | 408 | Y>F | No | Ensembl | |
| rs778846333 | 408 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1039875189 | 409 | S>Y | No | Ensembl | |
| rs1189153117 | 410 | F>I | No |
TOPMed gnomAD |
|
| rs1189153117 | 410 | F>L | No |
TOPMed gnomAD |
|
| rs1391885924 | 412 | I>F | No | gnomAD | |
| rs758525658 | 412 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs780221031 | 413 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs780221031 | 413 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1274680269 | 414 | A>V | No |
TOPMed gnomAD |
|
| rs539448650 | 415 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs768670915 | 416 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1467309835 | 416 | N>S | No |
TOPMed gnomAD |
|
|
COSM1205488 rs557614302 COSM1205487 |
418 | V>M | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes TOPMed gnomAD |
| rs543209822 | 420 | D>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs543209822 | 420 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1557572372 | 421 | L>P | No | Ensembl | |
| rs1641304960 | 422 | S>G | No | TOPMed | |
| rs1219125096 | 423 | P>R | No | gnomAD | |
| rs879209686 | 424 | E>A | No | Ensembl | |
| rs759275181 | 424 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs759275181 | 424 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1326774605 | 426 | R>C | No |
TOPMed gnomAD |
|
| rs1326774605 | 426 | R>G | No |
TOPMed gnomAD |
|
| rs767109730 | 426 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767109730 | 426 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs775160955 | 427 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs370118102 | 427 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs370118102 | 427 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs775160955 | 427 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2148253472 | 428 | A>S | No | Ensembl | |
| rs765172041 | 429 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs758170369 | 430 | V>G | No |
ExAC gnomAD |
|
| rs1458128053 | 431 | V>A | No | gnomAD | |
| rs1458128053 | 431 | V>G | No | gnomAD | |
| rs1208835088 | 431 | V>I | No | TOPMed | |
| rs150196251 | 432 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1436470617 | 433 | I>N | No | gnomAD | |
| rs1436470617 | 433 | I>T | No | gnomAD | |
| rs755050279 | 433 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4927537 COSM4927536 |
434 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201286524 | 435 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1019481620 | 436 | N>S | No |
TOPMed gnomAD |
|
| rs1019481620 | 436 | N>T | No |
TOPMed gnomAD |
|
| rs1016134236 | 437 | Q>H | No | Ensembl | |
| rs1330123469 | 438 | A>S | No |
TOPMed gnomAD |
|
| rs1330123469 | 438 | A>T | No |
TOPMed gnomAD |
|
| rs780557192 | 439 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs780557192 | 439 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs970788472 | 440 | P>L | No |
TOPMed gnomAD |
|
| rs970788472 | 440 | P>R | No |
TOPMed gnomAD |
|
| rs1233277139 | 440 | P>S | No | gnomAD | |
| rs1227374950 | 441 | S>F | No | gnomAD | |
| rs1641423318 | 442 | Q>E | No | TOPMed | |
| rs1269226681 | 442 | Q>R | No | gnomAD | |
| rs1641423636 | 444 | V>A | No | TOPMed | |
|
rs2295021 VAR_022107 |
444 | V>M | No |
UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs770292834 | 447 | R>C | No |
ExAC gnomAD |
|
| rs770292834 | 447 | R>G | No |
ExAC gnomAD |
|
| rs774280016 | 447 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1641424230 | 448 | Q>R | No | TOPMed | |
| rs1364164366 | 450 | R>G | No | gnomAD | |
| rs374661974 | 450 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM905219 rs1364164366 |
450 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1641424777 | 451 | A>T | No |
TOPMed gnomAD |
|
| rs775244487 | 451 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1641425184 | 453 | Q>E | No | Ensembl | |
| rs1570018365 | 454 | T>A | No | Ensembl | |
| rs1570018365 | 454 | T>P | No | Ensembl | |
| rs1304483173 | 455 | S>G | No | gnomAD | |
| rs1557576133 | 455 | S>I | No | Ensembl | |
| rs374046323 | 456 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374046323 | 456 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1288519265 | 457 | S>L | No |
TOPMed gnomAD |
|
| rs1288519265 | 457 | S>W | No |
TOPMed gnomAD |
|
| rs867788111 | 459 | L>M | No | gnomAD | |
| rs761966259 | 461 | Q>H | No |
ExAC TOPMed |
|
| rs1243010815 | 461 | Q>R | No |
TOPMed gnomAD |
|
| rs77608596 | 462 | E>G | No |
ExAC gnomAD |
|
| rs765337661 | 462 | E>K | No | ExAC | |
| rs758958503 | 463 | P>A | No |
ExAC gnomAD |
|
| rs1641426812 | 463 | P>L | No | Ensembl | |
| rs758958503 | 463 | P>S | No |
ExAC gnomAD |
|
| rs777499719 | 464 | E>G | No | ExAC | |
| rs755252711 | 464 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs755252711 | 464 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs748829132 | 465 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs778380990 | 466 | P>L | No |
ExAC gnomAD |
|
| rs778380990 | 466 | P>Q | No |
ExAC gnomAD |
|
| rs778380990 | 466 | P>R | No |
ExAC gnomAD |
|
| rs756827993 | 466 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs771968074 | 467 | N>S | No |
ExAC gnomAD |
|
| rs932518395 | 468 | G>S | No | Ensembl | |
| rs746841294 | 469 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1425666628 | 470 | I>T | No | gnomAD | |
| TCGA novel | 472 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1425283677 | 472 | E>Q | No | gnomAD | |
| rs768277316 | 473 | Y>* | No |
ExAC gnomAD |
|
| rs1641428976 | 474 | E>* | No |
TOPMed gnomAD |
|
| rs1641429067 | 474 | E>V | No |
TOPMed gnomAD |
|
| rs1641429204 | 475 | I>N | No | TOPMed | |
| rs1641429272 | 476 | K>M | No | TOPMed | |
| TCGA novel | 478 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761932406 | 479 | E>K | No |
ExAC TOPMed gnomAD |
|
| COSM905220 | 483 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 483 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1409847300 | 483 | E>K | No |
TOPMed gnomAD |
|
| rs1289866171 | 483 | E>V | No |
TOPMed gnomAD |
|
| rs751569232 | 484 | M>I | No |
ExAC gnomAD |
|
| rs779424856 | 484 | M>R | No |
ExAC gnomAD |
|
| rs199567884 | 485 | Q>E | No | 1000Genomes | |
| rs754845150 | 485 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs781079148 | 486 | S>R | No |
ExAC gnomAD |
|
| rs142120877 | 486 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1641433435 | 487 | Y>* | No | TOPMed | |
| rs1641433315 | 487 | Y>H | No | gnomAD | |
| rs1302396548 | 489 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs563025239 | 489 | T>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1641433783 | 490 | L>F | No | TOPMed | |
| rs149335835 | 491 | K>R | No |
ESP ExAC |
|
| rs1557576512 | 492 | A>T | No | Ensembl | |
|
TCGA novel rs1641434044 |
492 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs143432921 | 493 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2148259984 | 494 | T>I | No | Ensembl | |
| rs962345382 | 499 | V>I | No |
TOPMed gnomAD |
|
| TCGA novel | 500 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746428281 | 501 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs746428281 | 501 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs772488857 | 503 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs772488857 | 503 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs761039847 | 504 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1365604075 | 504 | P>S | No | gnomAD | |
| rs1325977423 | 505 | G>S | No | gnomAD | |
| rs1641435042 | 506 | T>I | No | Ensembl | |
|
rs182997024 COSM4029300 |
507 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs182997024 | 507 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1328139113 | 507 | R>H | No |
TOPMed gnomAD |
|
| rs182997024 | 507 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs55964751 | 508 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs955485235 | 508 | Y>C | No | Ensembl | |
| rs767371482 | 509 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs767371482 | 509 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1368378008 | 510 | F>L | No | TOPMed | |
| rs2148260104 | 511 | Q>H | No | 1000Genomes | |
| rs756044795 | 512 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs777700778 | 513 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs552762462 | 513 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1481247681 | 514 | A>T | No | gnomAD | |
| rs1156503379 | 514 | A>V | No | gnomAD | |
| rs757390422 | 515 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs757390422 | 515 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs779157641 | 515 | R>H | No |
ExAC gnomAD |
|
| rs1176745302 | 516 | T>A | No | gnomAD | |
| rs1436355220 | 517 | S>L | No | gnomAD | |
| rs1303892334 | 519 | G>D | No |
TOPMed gnomAD |
|
| rs1303892334 | 519 | G>V | No |
TOPMed gnomAD |
|
| rs772130793 | 520 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1408235070 | 522 | R>C | No |
TOPMed gnomAD |
|
| rs1408235070 | 522 | R>G | No |
TOPMed gnomAD |
|
|
COSM1205485 rs148500309 |
522 | R>H | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1408235070 | 522 | R>S | No |
TOPMed gnomAD |
|
| rs1272570562 | 525 | Q>* | No |
TOPMed gnomAD |
|
| rs769067119 | 525 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs149084883 | 525 | Q>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1641438176 | 526 | A>T | No | TOPMed | |
| rs1319904657 | 526 | A>V | No | gnomAD | |
|
TCGA novel rs762703677 |
527 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC TOPMed gnomAD |
| rs776953716 | 527 | M>T | No |
ExAC gnomAD |
|
| rs1220724691 | 527 | M>V | No | gnomAD | |
| rs1641438599 | 529 | V>L | No | Ensembl | |
| rs560791662 | 532 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771134902 | 533 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs1053441876 | 534 | P>T | No |
TOPMed gnomAD |
|
| rs1327893763 | 535 | R>Q | No |
TOPMed gnomAD |
|
| rs753717948 | 535 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs139374448 | 536 | P>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs946665358 | 536 | P>H | No |
TOPMed gnomAD |
|
| rs764034102 | 537 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs61751024 | 537 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs61751024 | 537 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs893984706 | 538 | Y>C | No |
TOPMed gnomAD |
|
| rs780142938 | 538 | Y>D | No |
ExAC TOPMed gnomAD |
|
| rs780142938 | 538 | Y>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 539 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1641484851 | 540 | T>I | No | TOPMed | |
| rs1641484922 | 541 | R>G | No | TOPMed | |
| TCGA novel | 542 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1223729255 | 542 | T>I | No |
TOPMed gnomAD |
|
| rs1223729255 | 542 | T>N | No |
TOPMed gnomAD |
|
| rs939191026 | 542 | T>S | No |
TOPMed gnomAD |
|
| rs751666192 | 543 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1401461753 | 543 | I>V | No |
TOPMed gnomAD |
|
|
rs754998997 COSM1667238 |
545 | W>* | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs754998997 | 545 | W>C | No |
ExAC TOPMed gnomAD |
|
| rs1009706519 | 546 | I>M | No |
TOPMed gnomAD |
|
| rs1557578304 | 548 | L>P | No | gnomAD | |
| rs138090615 | 549 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1196903163 | 550 | L>F | No |
TOPMed gnomAD |
|
| rs1196903163 | 550 | L>V | No |
TOPMed gnomAD |
|
| rs770117043 | 552 | T>A | No |
ExAC gnomAD |
|
| rs778181369 | 552 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs778181369 | 552 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs745551161 | 553 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs202042511 | 555 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202042511 | 555 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs760276645 | 556 | V>L | No |
ExAC gnomAD |
|
| rs1321075335 | 557 | L>I | No | gnomAD | |
| rs1641487298 | 557 | L>P | No | TOPMed | |
| rs1641487456 | 558 | L>P | No | TOPMed | |
| rs200214765 | 559 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1641487714 | 560 | L>P | No | Ensembl | |
| rs973512254 | 562 | I>V | No | TOPMed | |
| rs1641488103 | 563 | C>F | No | gnomAD | |
| rs776497408 | 563 | C>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 563 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1641488269 | 564 | K>E | No | Ensembl | |
| rs1641488348 | 564 | K>R | No | TOPMed | |
| TCGA novel | 566 | R>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759735882 | 567 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs774616715 | 567 | H>Y | No |
ExAC gnomAD |
|
| COSM3789612 | 568 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767538683 | 568 | C>Y | No |
ExAC gnomAD |
|
| rs1164990116 | 571 | S>N | No | gnomAD | |
| COSM4029301 | 573 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1298332967 | 574 | F>C | No |
TOPMed gnomAD |
|
| rs756098365 | 574 | F>V | No |
ExAC TOPMed gnomAD |
|
| rs764676638 | 576 | D>N | No |
ExAC gnomAD |
|
| rs754330833 | 577 | S>L | No |
ExAC gnomAD |
|
| rs1641511092 | 577 | S>P | No | Ensembl | |
| rs370682222 | 578 | D>E | No |
ESP ExAC gnomAD |
|
| rs531926893 | 579 | E>K | No |
ExAC gnomAD |
|
| rs531926893 | 579 | E>Q | No |
ExAC gnomAD |
|
| rs754645453 | 581 | K>Q | No |
ExAC gnomAD |
|
| rs138590921 | 581 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs138590921 | 581 | K>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374833210 | 582 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs769125415 | 582 | M>T | No | ExAC | |
| rs901903397 | 583 | H>Q | No |
TOPMed gnomAD |
|
| rs773202551 | 583 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs2148263800 | 584 | Y>C | No | Ensembl | |
| rs749212078 | 585 | Q>H | No |
ExAC gnomAD |
|
| rs376737470 | 585 | Q>P | No |
TOPMed gnomAD |
|
| rs376737470 | 585 | Q>R | No |
TOPMed gnomAD |
|
| COSM3864716 | 587 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1200501191 | 588 | Q>E | No | gnomAD | |
| rs774104397 | 589 | A>T | No |
ExAC gnomAD |
|
| rs761940802 | 589 | A>V | No |
ExAC gnomAD |
|
| rs1472161891 | 590 | P>H | No | TOPMed | |
| rs765867803 | 590 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1221309032 | 591 | P>Q | No | gnomAD | |
| rs1221309032 | 591 | P>R | No | gnomAD | |
| rs1641545402 | 595 | L>R | No | TOPMed | |
| TCGA novel | 597 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763379733 | 598 | H>R | No |
ExAC gnomAD |
|
| rs766646724 | 599 | H>L | No |
ExAC TOPMed |
|
| rs766646724 | 599 | H>R | No |
ExAC TOPMed |
|
| rs202243952 | 600 | P>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 600 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs202243952 | 600 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs753369755 | 601 | P>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 601 | P>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753369755 | 601 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1396986101 | 602 | G>E | No | gnomAD | |
| COSM3864717 | 603 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745792239 | 603 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1641546750 | 604 | L>F | No | Ensembl | |
| rs1367739182 | 604 | L>P | No | gnomAD | |
| rs1641547055 | 606 | E>D | No | Ensembl | |
| rs1641546990 | 606 | E>G | No | Ensembl | |
| rs144329757 | 607 | P>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs144329757 | 607 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs771963113 | 607 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1339995453 | 608 | Q>* | No | TOPMed | |
| rs1311825398 | 608 | Q>H | No | TOPMed | |
| rs141532503 | 608 | Q>L | No |
ESP ExAC TOPMed gnomAD |
|
|
TCGA novel rs768788331 |
609 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC gnomAD NCI-TCGA |
| rs1015313398 | 610 | Y>C | No | gnomAD | |
| rs776940252 | 611 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs776940252 | 611 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
VAR_024514 rs999765 |
612 | E>Q | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1483434281 | 613 | P>L | No | Ensembl | |
| rs763353844 | 613 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs766808717 | 614 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1570028542 | 615 | T>A | No | Ensembl | |
| rs2148265421 | 615 | T>N | No | Ensembl | |
| rs1232730345 | 616 | Y>H | No | gnomAD | |
| rs371894783 | 617 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1641549355 | 618 | E>D | No | TOPMed | |
| rs866840934 | 618 | E>K | No | Ensembl | |
| rs934020406 | 619 | P>L | No | TOPMed | |
| rs753426993 | 620 | G>V | No |
ExAC TOPMed gnomAD |
|
| COSM1284757 | 621 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756831723 | 621 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs756831723 | 621 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1164315190 | 621 | R>W | No |
TOPMed gnomAD |
|
| rs758235140 | 622 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1641550115 | 622 | A>V | No |
TOPMed gnomAD |
|
| rs1333072744 | 624 | R>C | No |
TOPMed gnomAD |
|
|
COSM239722 rs746799254 |
624 | R>H | Variant assessed as Somatic; MODERATE impact. prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs754761927 | 625 | S>G | No |
ExAC gnomAD |
|
| rs1357841927 | 627 | T>A | No |
TOPMed gnomAD |
|
| rs781568823 | 628 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs748375751 | 628 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs781568823 | 628 | R>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 629 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376201963 | 629 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs376201963 | 629 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs369284774 | 630 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1197301453 | 630 | I>S | No |
TOPMed gnomAD |
|
| rs771365576 | 631 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1461270305 | 632 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1239494708 | 633 | S>C | No | TOPMed | |
| COSM905221 | 634 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1641551876 | 636 | H>P | No | TOPMed | |
| rs774864430 | 637 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1184750288 | 637 | I>V | No | gnomAD | |
| rs567750244 | 638 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs567750244 | 638 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs373638442 | 641 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs536702821 | 642 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1641552595 | 643 | S>C | No | Ensembl | |
| rs1172572081 | 643 | S>P | No | gnomAD | |
| rs533721836 | 645 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs533721836 | 645 | D>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1641559376 | 646 | S>A | No | Ensembl | |
| rs1468602131 | 647 | G>R | No |
TOPMed gnomAD |
|
| rs199682979 | 649 | V>F | No | Ensembl | |
| rs757343485 | 650 | C>G | No |
ExAC TOPMed gnomAD |
|
| rs757343485 | 650 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1021905173 | 651 | Y>* | No |
TOPMed gnomAD |
|
| rs779031605 | 652 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs200774719 | 655 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs746416885 | 655 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1641560789 | 656 | V>L | No | TOPMed | |
| rs1334346160 | 657 | P>L | No | gnomAD | |
| rs1641560977 | 657 | P>S | No | gnomAD | |
| rs747383112 | 660 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs747383112 | 660 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1239030664 | 660 | R>W | No |
TOPMed gnomAD |
|
| COSM3750977 | 661 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772805814 | 661 | D>H | No |
ExAC gnomAD |
|
| rs762652898 | 661 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs770394863 | 662 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs770394863 | 662 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs960442770 | 664 | V>L | No |
TOPMed gnomAD |
|
| rs960442770 | 664 | V>M | No |
TOPMed gnomAD |
|
| rs368786183 | 666 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs752615573 | 666 | I>M | No |
ExAC gnomAD |
|
| rs2148266081 | 667 | K>E | No | Ensembl | |
| rs760614050 | 667 | K>T | No |
ExAC gnomAD |
|
| rs1387248062 | 669 | L>F | No | gnomAD | |
| COSM4029303 | 671 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757472636 | 672 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs757472636 | 672 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1641562733 | 672 | G>V | No | Ensembl | |
| rs1458154418 | 673 | Y>* | No | gnomAD | |
| rs750521094 | 674 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs750521094 | 674 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs373953943 | 675 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1339980 | 675 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs562829959 | 679 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs562829959 | 679 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs557674245 | 679 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1570030034 | 680 | D>G | No | Ensembl | |
| rs770640699 | 684 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1360246232 | 685 | A>S | No |
TOPMed gnomAD |
|
| TCGA novel | 685 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201485535 | 685 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1570030099 | 686 | S>A | No | Ensembl | |
| rs1570030099 | 686 | S>P | No | Ensembl | |
| rs1641564346 | 687 | I>T | No | TOPMed | |
| rs775630538 | 688 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs775630538 | 688 | M>V | No |
ExAC TOPMed gnomAD |
|
|
rs760643869 COSM905222 |
689 | G>R | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs2148266209 | 689 | G>V | No | Ensembl | |
| rs763837839 | 692 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs763837839 | 692 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1166522062 | 693 | H>R | No | gnomAD | |
| rs542047673 | 694 | P>S | No |
1000Genomes ExAC gnomAD |
|
| rs77067006 | 696 | I>T | No | Ensembl | |
| rs765456520 | 696 | I>V | No |
ExAC gnomAD |
|
| rs1641565476 | 697 | I>S | No | gnomAD | |
| rs141165537 | 698 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141165537 | 698 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3400446 rs758537106 |
698 | R>H | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs758537106 | 698 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs758537106 | 698 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs766433025 | 700 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs766433025 | 700 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1383674516 | 702 | V>F | No | gnomAD | |
| rs1383674516 | 702 | V>I | No | gnomAD | |
| rs755476945 | 703 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs528574353 | 705 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs528574353 | 705 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs140243523 | 705 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs140243523 | 705 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs769570525 | 706 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1258547318 | 706 | G>S | No | gnomAD | |
|
rs62642537 RCV000964803 |
707 | R>C | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs769624500 COSM3400447 |
707 | R>H | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1300070575 | 709 | A>T | No | gnomAD | |
| rs1363969002 | 709 | A>V | No | gnomAD | |
| rs1641576315 | 711 | I>F | No | Ensembl | |
| rs541620093 | 711 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149392840 | 712 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs771163012 | 714 | E>G | No |
ExAC gnomAD |
|
| rs1220926809 | 715 | Y>C | No |
TOPMed gnomAD |
|
| rs1220926809 | 715 | Y>F | No |
TOPMed gnomAD |
|
| rs1641576932 | 716 | M>V | No | TOPMed | |
| TCGA novel | 717 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146955063 | 719 | G>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs146955063 | 719 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1641577428 | 720 | S>F | No | TOPMed | |
| rs1641577347 | 720 | S>P | No |
TOPMed gnomAD |
|
| rs753297349 | 721 | L>P | No | ExAC | |
| COSM679200 | 722 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1570031346 | 723 | T>A | No | Ensembl | |
| rs1641577690 | 723 | T>I | No | Ensembl | |
| rs1570031346 | 723 | T>P | No | Ensembl | |
| rs1641577823 | 725 | L>P | No | TOPMed | |
| rs755562643 | 727 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs971991308 | 729 | D>N | No | gnomAD | |
| rs778542762 | 730 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1442302244 | 731 | Q>* | No | gnomAD | |
| rs1442302244 | 731 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1210934249 | 734 | I>M | No | gnomAD | |
| rs1231061944 | 734 | I>S | No | TOPMed | |
| rs1641595652 | 734 | I>V | No | TOPMed | |
| rs1641595988 | 735 | M>I | No | TOPMed | |
| rs200018942 | 735 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs980221016 | 736 | Q>E | No | gnomAD | |
| rs1641596386 | 736 | Q>H | No | TOPMed | |
| rs2148267710 | 736 | Q>P | No | Ensembl | |
| rs1570032915 | 738 | V>M | No | Ensembl | |
| rs1641596564 | 739 | G>S | No |
TOPMed gnomAD |
|
| rs371556187 | 740 | M>I | No |
ESP TOPMed gnomAD |
|
| rs1469581961 | 740 | M>T | No | TOPMed | |
| rs936854890 | 742 | R>K | No |
TOPMed gnomAD |
|
| rs138538362 | 743 | G>R | No |
ESP TOPMed gnomAD |
|
| rs1403843416 | 744 | V>A | No | gnomAD | |
| rs1175240210 | 744 | V>M | No | TOPMed | |
|
TCGA novel rs1641597315 |
746 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs200648943 | 747 | G>S | No |
ExAC gnomAD |
|
| rs1360380857 | 748 | M>T | No | gnomAD | |
| rs147240669 | 749 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs201964877 | 749 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1341561708 | 750 | Y>C | No | gnomAD | |
| rs552850440 | 751 | L>F | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 751 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs552850440 | 751 | L>V | No |
1000Genomes ExAC gnomAD |
|
| rs1051630007 | 752 | S>L | No | Ensembl | |
| rs763326604 | 753 | D>E | No |
ExAC gnomAD |
|
| rs767089293 | 754 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1570033099 | 754 | L>P | No | Ensembl | |
| rs767089293 | 754 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1641599161 | 755 | G>D | No |
TOPMed gnomAD |
|
| rs752279731 | 756 | Y>C | No |
ExAC gnomAD |
|
|
rs1641599587 COSM4029306 |
759 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs757176752 | 759 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2148267857 | 760 | D>H | No | Ensembl | |
| TCGA novel | 760 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1641599810 | 761 | L>P | No | Ensembl | |
| rs1641599886 | 762 | A>T | No | TOPMed | |
| rs149930866 | 763 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs149930866 | 763 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2148267884 | 763 | A>V | No | Ensembl | |
| rs531451396 | 764 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747216998 | 764 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs747216998 | 764 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs768903431 | 765 | N>I | No |
ExAC gnomAD |
|
| rs768903431 | 765 | N>S | No |
ExAC gnomAD |
|
| rs370473123 | 766 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs370473123 | 766 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs568027987 | 768 | V>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs568027987 | 768 | V>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs147123754 | 768 | V>I | No |
1000Genomes ExAC gnomAD |
|
| rs1641601177 | 769 | D>G | No | gnomAD | |
| rs1277091018 | 770 | S>N | No | gnomAD | |
| rs1257012529 | 771 | N>K | No | gnomAD | |
| TCGA novel | 771 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1641601631 | 775 | K>N | No | TOPMed | |
| rs1239542886 | 776 | V>M | No | gnomAD | |
| rs1165586714 | 777 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1165586714 | 777 | S>F | No |
TOPMed gnomAD |
|
| rs1557582337 | 779 | F>I | No | Ensembl | |
| rs1641602497 | 780 | G>E | No | Ensembl | |
| rs753371979 | 780 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs750373572 | 783 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM1339981 rs750373572 |
783 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs202070589 | 783 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1641603118 | 784 | V>M | No | TOPMed | |
| rs779969785 | 787 | D>A | No | ExAC | |
| rs779969785 | 787 | D>G | No | ExAC | |
| rs758182002 | 787 | D>Y | No |
ExAC gnomAD |
|
| rs1641603648 | 788 | D>G | No | TOPMed | |
| rs755279880 | 788 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1641603782 | 789 | P>A | No | gnomAD | |
| rs369475595 | 789 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
|
TCGA novel rs2148268075 |
790 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1334522181 | 792 | A>S | No |
TOPMed gnomAD |
|
| TCGA novel | 793 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1436163411 | 794 | T>A | No | gnomAD | |
| TCGA novel | 794 | T>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1641604711 | 794 | T>I | No | gnomAD | |
| rs771326608 | 795 | T>A | No |
ExAC gnomAD |
|
| rs774731033 | 796 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs774731033 | 796 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs537363813 | 797 | G>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs150954894 | 797 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs758807841 | 798 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs780431554 | 799 | K>E | No |
ExAC gnomAD |
|
| rs1570039820 | 800 | I>N | No | TOPMed | |
| rs1570039820 | 800 | I>T | No | TOPMed | |
| rs769507116 | 803 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs368283503 | 803 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs368283503 | 803 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs372280342 COSM1339982 |
805 | T>M | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1175953678 | 809 | A>T | No | gnomAD | |
| rs915112154 | 810 | I>F | No |
TOPMed gnomAD |
|
| rs555665976 | 811 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1047491791 | 811 | A>V | No | Ensembl | |
| rs775378376 | 813 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs760332050 | 813 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs760332050 | 813 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1390787117 | 815 | F>L | No |
TOPMed gnomAD |
|
| rs1300462328 | 816 | S>F | No | gnomAD | |
| rs757340105 | 817 | S>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755230170 | 817 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs374741939 | 817 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3484900 rs939029576 |
818 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs750964170 | 820 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs758930799 | 820 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1041225432 | 821 | V>A | No | Ensembl | |
| rs747411921 | 821 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs747411921 | 821 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1641699890 | 822 | W>C | No | TOPMed | |
|
COSM3400448 rs1641699821 |
822 | W>R | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1641700202 | 825 | G>D | No | TOPMed | |
| rs369148933 | 825 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs72651352 | 826 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1346788152 | 828 | M>I | No | gnomAD | |
| rs1160657874 | 828 | M>T | No |
TOPMed gnomAD |
|
| rs1641700702 | 829 | W>* | No | TOPMed | |
|
rs1456223293 COSM1756864 |
830 | E>K | urinary_tract [Cosmic] | No |
cosmic curated gnomAD |
| rs1368228241 | 831 | V>M | No | gnomAD | |
| rs1426560003 | 833 | A>G | No | TOPMed | |
| COSM4029308 | 833 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1570040341 | 834 | Y>C | No | Ensembl | |
| rs144705335 | 837 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201817606 | 837 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372238663 | 838 | P>L | No |
ESP TOPMed gnomAD |
|
| rs1162167276 | 839 | Y>H | No |
TOPMed gnomAD |
|
| rs1162167276 | 839 | Y>N | No |
TOPMed gnomAD |
|
|
TCGA novel rs768326430 |
840 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
| rs2148272592 | 844 | N>Y | No | Ensembl | |
| rs761988321 | 845 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs761988321 | 845 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs569320402 COSM905225 |
845 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1641706026 | 848 | I>V | No | gnomAD | |
| rs1009623594 | 849 | S>T | No |
TOPMed gnomAD |
|
| rs1570040780 | 850 | S>F | No | TOPMed | |
| rs1216823619 | 851 | V>M | No | gnomAD | |
| rs1441885608 | 853 | E>D | No | gnomAD | |
| COSM679199 | 855 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1570040814 | 855 | Y>D | No | TOPMed | |
|
rs1570040814 COSM1559284 |
855 | Y>N | central_nervous_system [Cosmic] | No |
cosmic curated TOPMed |
| rs146743212 | 856 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV000785719 rs149877319 |
856 | R>H | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs149877319 | 856 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2148272841 | 857 | L>M | No | Ensembl | |
| rs1336826933 | 857 | L>Q | No | gnomAD | |
|
rs1191247338 COSM3864718 |
858 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs749447607 | 858 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs200796590 | 859 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200796590 | 859 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374551936 | 860 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| VAR_042158 | 860 | P>L | a metastatic melanoma sample; somatic mutation [UniProt] | No | UniProt |
|
rs374551936 COSM1339983 |
860 | P>S | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs374551936 | 860 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs35887233 | 861 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs776137702 | 861 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs567663699 | 861 | M>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776137702 | 861 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs367752791 | 865 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1183460086 | 865 | H>Y | No |
TOPMed gnomAD |
|
| rs766004176 | 866 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1235385455 | 866 | A>V | No |
TOPMed gnomAD |
|
| rs1641708541 | 867 | L>P | No | Ensembl | |
| COSM6124671 | 868 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751041267 | 868 | H>Y | No |
ExAC gnomAD |
|
| rs1282486492 | 871 | M>I | No | gnomAD | |
| rs781170423 | 871 | M>R | No |
ExAC gnomAD |
|
| rs781170423 | 871 | M>T | No |
ExAC gnomAD |
|
| rs1641708978 | 872 | L>V | No | TOPMed | |
| rs1472297861 | 873 | D>A | No | gnomAD | |
| rs755951383 | 873 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs777512975 | 874 | C>S | No |
ExAC gnomAD |
|
| rs749071269 | 875 | W>* | No |
ExAC gnomAD |
|
| rs1307708916 | 875 | W>G | No | TOPMed | |
| rs1164998225 | 876 | H>P | No | gnomAD | |
| rs950616882 | 877 | K>E | No | TOPMed | |
| rs1195008074 | 877 | K>R | No |
TOPMed gnomAD |
|
| rs771071426 | 878 | D>G | No |
ExAC gnomAD |
|
| TCGA novel | 879 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146856523 | 879 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146856523 | 879 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147803148 | 879 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs761229579 | 880 | A>T | No |
ExAC TOPMed gnomAD |
|
|
rs764652650 COSM1743271 |
880 | A>V | biliary_tract Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1402831345 | 881 | Q>P | No |
TOPMed gnomAD |
|
| rs765954947 | 882 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2148273057 | 882 | R>W | No | Ensembl | |
| rs1331042657 | 883 | P>R | No | gnomAD | |
| rs751222767 | 884 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs751222767 | 884 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs62618734 RCV000974592 COSM1559286 |
884 | R>H | central_nervous_system [Cosmic] | No |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1641711401 | 886 | S>A | No | gnomAD | |
| rs1374737604 | 886 | S>F | No | gnomAD | |
| rs1641711604 | 887 | Q>E | No | Ensembl | |
| rs1353049777 | 888 | I>T | No |
TOPMed gnomAD |
|
| rs1287388136 | 892 | L>F | No | gnomAD | |
| rs1254387084 | 893 | D>G | No |
TOPMed gnomAD |
|
| rs544797559 | 893 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs138872731 | 894 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1441761932 | 895 | L>F | No | TOPMed | |
| rs375664082 | 896 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs375664082 | 896 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs146256137 | 897 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772315293 | 897 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs772315293 | 897 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs780307964 | 898 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1398683463 | 899 | P>R | No | gnomAD | |
| rs747032878 | 899 | P>S | No |
ExAC gnomAD |
|
| rs1309992168 | 902 | L>F | No | gnomAD | |
| rs200152015 | 905 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs200152015 | 905 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs142726625 | 906 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs142726625 | 906 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1214049387 | 906 | A>V | No | gnomAD | |
| rs1641713845 | 908 | V>D | No | gnomAD | |
| rs1456340962 | 909 | S>N | No | gnomAD | |
| rs1197464122 | 909 | S>R | No | gnomAD | |
| rs1641714286 | 910 | R>M | No | TOPMed | |
| rs1641719763 | 911 | C>Y | No | Ensembl | |
| TCGA novel | 912 | P>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748668673 | 912 | P>L | No |
ExAC gnomAD |
|
| rs199853592 | 913 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs199853592 | 913 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs571732433 | 913 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763255096 | 914 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1641720185 | 914 | P>S | No | gnomAD | |
| rs1190514725 | 915 | A>T | No |
TOPMed gnomAD |
|
| rs2148273587 | 917 | V>A | No | Ensembl | |
| rs147795823 | 917 | V>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147795823 | 917 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147795823 | 917 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs141279306 COSM4029310 |
918 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs760234665 | 918 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs753756295 | 922 | D>G | No |
ExAC gnomAD |
|
| rs1043147569 | 922 | D>N | No | TOPMed | |
| rs778640305 | 923 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs765031953 | 924 | R>* | No |
ExAC gnomAD |
|
| rs147939357 | 924 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147939357 | 924 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147939357 | 924 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201047906 | 925 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs200470501 | 925 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1641721848 | 926 | G>C | No |
TOPMed gnomAD |
|
| rs143804645 | 926 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1641722001 | 927 | S>I | No | TOPMed | |
| rs1641722001 | 927 | S>N | No | TOPMed | |
| rs781303625 | 928 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1049095478 | 929 | G>A | No | gnomAD | |
| rs1049095478 | 929 | G>D | No | gnomAD | |
|
COSM905226 rs756668263 |
930 | G>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1488149108 | 931 | G>E | No | gnomAD | |
| rs142066703 | 932 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs778194934 | 932 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs778194934 | 932 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs142066703 | 932 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1431878746 | 934 | T>P | No | gnomAD | |
| rs1198331977 | 934 | T>S | No | gnomAD | |
| rs1019169249 | 935 | V>M | No |
TOPMed gnomAD |
|
| rs746640832 | 936 | G>R | No |
ExAC gnomAD |
|
| rs1289170642 | 937 | D>N | No | gnomAD | |
| rs776224376 | 938 | W>* | No |
ExAC gnomAD |
|
| rs765228796 | 939 | L>R | No | ExAC | |
| rs758336010 | 939 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1641724022 | 940 | D>A | No | Ensembl | |
| rs1641724022 | 940 | D>G | No | Ensembl | |
|
rs374945795 COSM331176 |
940 | D>H | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs374945795 | 940 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs199559772 | 943 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs755260283 COSM1251353 |
943 | R>H | oesophagus [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs755260283 | 943 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs199559772 | 943 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3484903 | 944 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM330173 rs1216966524 |
945 | G>D | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
rs146237497 COSM1205484 |
946 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs752835457 | 946 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs749771471 | 948 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs149515751 | 948 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs565216451 | 949 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1253321212 | 950 | H>N | No |
TOPMed gnomAD |
|
|
rs746166476 COSM3385877 |
952 | A>T | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed |
| rs1641725579 | 952 | A>V | No | TOPMed | |
| rs369076646 | 953 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs1468847722 | 954 | G>S | No | TOPMed | |
|
COSM293374 rs990213044 |
955 | G>R | Variant assessed as Somatic; MODERATE impact. oesophagus large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1325102343 | 956 | Y>C | No |
TOPMed gnomAD |
|
| rs912923867 | 957 | S>F | No | TOPMed | |
| COSM6124669 | 958 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1641726401 | 958 | S>Y | No | TOPMed | |
| TCGA novel | 959 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs576952185 | 959 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs747764837 | 959 | L>V | No |
ExAC gnomAD |
|
| rs762954946 | 961 | M>I | No |
ExAC gnomAD |
|
| rs774311718 | 964 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs774311718 | 964 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs202016361 | 964 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs202016361 | 964 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1394659148 | 965 | M>T | No | TOPMed | |
| rs752849070 | 965 | M>V | No |
ExAC gnomAD |
|
| rs373032976 | 966 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1641728004 | 966 | N>S | No | Ensembl | |
| rs375902715 | 967 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1237632422 | 968 | Q>L | No | gnomAD | |
| TCGA novel | 969 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs148692719 | 970 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148692719 | 970 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776890427 | 971 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs139543017 | 971 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs139543017 | 971 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2148274373 | 972 | A>D | No | Ensembl | |
| TCGA novel | 972 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs145095203 | 972 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145095203 | 972 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1273206543 | 974 | G>D | No |
TOPMed gnomAD |
|
| rs1235063910 | 976 | T>N | No | gnomAD | |
| rs1305930113 | 977 | L>H | No | gnomAD | |
| rs766802239 | 978 | M>I | No |
ExAC gnomAD |
|
| rs536376846 | 978 | M>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs958885328 | 978 | M>R | No |
TOPMed gnomAD |
|
| rs536376846 | 978 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1447568896 | 979 | G>S | No | gnomAD | |
| rs1641739156 | 980 | H>Y | No | Ensembl | |
| rs1217829614 | 982 | K>E | No | gnomAD | |
| rs1250639728 | 982 | K>N | No |
TOPMed gnomAD |
|
| rs1217829614 | 982 | K>Q | No | gnomAD | |
| rs199735359 | 983 | K>M | No |
ExAC TOPMed gnomAD |
|
| rs1396520336 | 984 | I>V | No |
TOPMed gnomAD |
|
| rs760037386 | 985 | L>P | No |
ExAC gnomAD |
|
| rs1402994229 | 986 | G>S | No | gnomAD | |
| rs2148274484 | 987 | S>R | No | Ensembl | |
| rs1641740065 | 987 | S>T | No | Ensembl | |
| rs767961686 | 989 | Q>* | No |
ExAC gnomAD |
|
| rs767961686 | 989 | Q>E | No |
ExAC gnomAD |
|
| rs1434042206 | 990 | T>S | No | TOPMed | |
| rs1327935510 | 991 | M>I | No | gnomAD | |
| rs756880433 | 991 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs199804540 | 992 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs199804540 | 992 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs542943867 COSM1205489 |
992 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed |
| rs200111539 | 993 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200111539 | 993 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs150038826 | 993 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs754811830 | 998 | T>I | No |
ExAC gnomAD |
|
| rs754811830 | 998 | T>S | No |
ExAC gnomAD |
|
| rs1570043585 | 999 | Q>* | No | Ensembl | |
| rs1011710043 | 1000 | G>R | No |
TOPMed gnomAD |
|
| rs1450177211 | 1001 | P>R | No |
TOPMed gnomAD |
|
| rs1275624041 | 1001 | P>S | No |
TOPMed gnomAD |
|
| rs781000303 | 1002 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs566456855 | 1002 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs370302532 | 1003 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374170042 | 1003 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs370302532 COSM1236217 |
1003 | R>W | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs1035348096 | 1005 | L>H | No | TOPMed | |
| rs200304246 | 1005 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
No associated diseases with P29322
6 regional properties for P29322
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 706 - 834 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 315 - 467 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 589 - 705 | IPR001711 |
| domain | Phospholipase C-beta, C-terminal domain | 943 - 1200 | IPR014815 |
| domain | PLC-beta, PH domain | 18 - 147 | IPR037862 |
| domain | Phosphoinositide phospholipase C beta 1-4-like, EF-hand domain | 152 - 219 | IPR053945 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ephrin receptor activity | Combining with an ephrin receptor ligand to initiate a change in cell activity. |
| GPI-linked ephrin receptor activity | Combining with a GPI-anchored ephrin to initiate a change in cell activity. |
| growth factor binding | Binding to a growth factor, proteins or polypeptides that stimulate a cell or organism to grow or proliferate. |
| transmembrane-ephrin receptor activity | Combining with a transmembrane ephrin to initiate a change in cell activity. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| cellular response to follicle-stimulating hormone stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a follicle-stimulating hormone stimulus. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| neuron projection development | The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| neuron remodeling | The developmentally regulated remodeling of neuronal projections such as pruning to eliminate the extra dendrites and axons projections set up in early stages of nervous system development. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| regulation of cell adhesion | Any process that modulates the frequency, rate or extent of attachment of a cell to another cell or to the extracellular matrix. |
| regulation of cell adhesion mediated by integrin | Any process that modulates the frequency, rate, or extent of cell adhesion mediated by integrin. |
| substrate-dependent cell migration | The orderly movement of a cell from one site to another along a substrate such as the extracellular matrix; the migrating cell forms a protrusion that attaches to the substrate. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P21709 | EPHA1 | Ephrin type-A receptor 1 | Homo sapiens (Human) | SS |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| Q15375 | EPHA7 | Ephrin type-A receptor 7 | Homo sapiens (Human) | SS |
| P54756 | EPHA5 | Ephrin type-A receptor 5 | Homo sapiens (Human) | SS |
| Q9UF33 | EPHA6 | Ephrin type-A receptor 6 | Homo sapiens (Human) | SS |
| P54762 | EPHB1 | Ephrin type-B receptor 1 | Homo sapiens (Human) | SS |
| P29320 | EPHA3 | Ephrin type-A receptor 3 | Homo sapiens (Human) | PR |
| P54764 | EPHA4 | Ephrin type-A receptor 4 | Homo sapiens (Human) | SS |
| P54753 | EPHB3 | Ephrin type-B receptor 3 | Homo sapiens (Human) | SS |
| Q5JZY3 | EPHA10 | Ephrin type-A receptor 10 | Homo sapiens (Human) | SS |
| O15197 | EPHB6 | Ephrin type-B receptor 6 | Homo sapiens (Human) | SS |
| P29323 | EPHB2 | Ephrin type-B receptor 2 | Homo sapiens (Human) | EV |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| O08644 | Ephb6 | Ephrin type-B receptor 6 | Mus musculus (Mouse) | PR |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| O61460 | vab-1 | Ephrin receptor 1 | Caenorhabditis elegans | SS |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E991 | PRK6 | Pollen receptor-like kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FXF2 | RKF1 | Probable LRR receptor-like serine/threonine-protein kinase RFK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O13147 | ephb3 | Ephrin type-B receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73878 | ephb4b | Ephrin type-B receptor 4b | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAPARGRLPP | ALWVVTAAAA | AATCVSAARG | EVNLLDTSTI | HGDWGWLTYP | AHGWDSINEV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DESFQPIHTY | QVCNVMSPNQ | NNWLRTSWVP | RDGARRVYAE | IKFTLRDCNS | MPGVLGTCKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TFNLYYLESD | RDLGASTQES | QFLKIDTIAA | DESFTGADLG | VRRLKLNTEV | RSVGPLSKRG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FYLAFQDIGA | CLAILSLRIY | YKKCPAMVRN | LAAFSEAVTG | ADSSSLVEVR | GQCVRHSEER |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DTPKMYCSAE | GEWLVPIGKC | VCSAGYEERR | DACVACELGF | YKSAPGDQLC | ARCPPHSHSA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| APAAQACHCD | LSYYRAALDP | PSSACTRPPS | APVNLISSVN | GTSVTLEWAP | PLDPGGRSDI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TYNAVCRRCP | WALSRCEACG | SGTRFVPQQT | SLVQASLLVA | NLLAHMNYSF | WIEAVNGVSD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LSPEPRRAAV | VNITTNQAAP | SQVVVIRQER | AGQTSVSLLW | QEPEQPNGII | LEYEIKYYEK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DKEMQSYSTL | KAVTTRATVS | GLKPGTRYVF | QVRARTSAGC | GRFSQAMEVE | TGKPRPRYDT |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RTIVWICLTL | ITGLVVLLLL | LICKKRHCGY | SKAFQDSDEE | KMHYQNGQAP | PPVFLPLHHP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PGKLPEPQFY | AEPHTYEEPG | RAGRSFTREI | EASRIHIEKI | IGSGDSGEVC | YGRLRVPGQR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DVPVAIKALK | AGYTERQRRD | FLSEASIMGQ | FDHPNIIRLE | GVVTRGRLAM | IVTEYMENGS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| LDTFLRTHDG | QFTIMQLVGM | LRGVGAGMRY | LSDLGYVHRD | LAARNVLVDS | NLVCKVSDFG |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LSRVLEDDPD | AAYTTTGGKI | PIRWTAPEAI | AFRTFSSASD | VWSFGVVMWE | VLAYGERPYW |
| 850 | 860 | 870 | 880 | 890 | 900 |
| NMTNRDVISS | VEEGYRLPAP | MGCPHALHQL | MLDCWHKDRA | QRPRFSQIVS | VLDALIRSPE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| SLRATATVSR | CPPPAFVRSC | FDLRGGSGGG | GGLTVGDWLD | SIRMGRYRDH | FAAGGYSSLG |
| 970 | 980 | 990 | 1000 | ||
| MVLRMNAQDV | RALGITLMGH | QKKILGSIQT | MRAQLTSTQG | PRRHL |