P29294
Gene name |
MYLK |
Protein name |
Myosin light chain kinase, smooth muscle |
Names |
MLCK, smMLCK, Telokin |
Species |
Oryctolagus cuniculus (Rabbit) |
KEGG Pathway |
ocu:100009143 |
EC number |
|
Protein Class |
IMMUNOGLOBULIN (PTHR47633) |
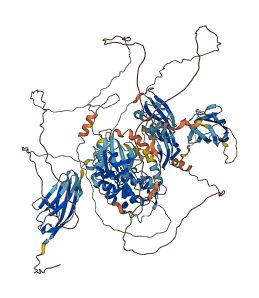
Descriptions
Myosin light chain kinases (MLCK) are members of the family of Ca2+-calmodulin-dependent protein kinases. The autoinhibitory pseudosubstrate sequence was identified by sequence similarity with the chicken smooth muscle MLCK (P11799-2). The mutational changes of acidic residues in the catalytic core of the kinase domain, which may interact with the basic residues in the pseudosubstrate sequence, increased the catalytic activity of the kinase domain.
Autoinhibitory domains (AIDs)
Target domain |
696-951 (Protein kinase domain) |
Relief mechanism |
Partner binding |
Assay |
Mutagenesis experiment, Deletion assay |
Accessory elements
836-858 (Activation loop from InterPro)
Target domain |
696-951 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Gallagher PJ et al. (1993) "A molecular mechanism for autoinhibition of myosin light chain kinases", The Journal of biological chemistry, 268, 26578-82
- Padre RC et al. (2000) "Functional assembly of fragments from bisected smooth muscle myosin light chain kinase", The Journal of biological chemistry, 275, 26665-73
Autoinhibited structure

Activated structure

1 structures for P29294
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P29294-F1 | Predicted | AlphaFoldDB |
No variants for P29294
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P29294 | |||||
No associated diseases with P29294
11 regional properties for P29294
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 696 - 951 | IPR000719 |
| domain | Immunoglobulin subtype | 335 - 419 | IPR003599-1 |
| domain | Immunoglobulin subtype | 475 - 559 | IPR003599-2 |
| domain | Immunoglobulin subtype | 1047 - 1132 | IPR003599-3 |
| domain | Fibronectin type III | 562 - 657 | IPR003961 |
| domain | Immunoglobulin-like domain | 329 - 417 | IPR007110-1 |
| domain | Immunoglobulin-like domain | 469 - 557 | IPR007110-2 |
| domain | Immunoglobulin-like domain | 1041 - 1130 | IPR007110-3 |
| active_site | Serine/threonine-protein kinase, active site | 813 - 825 | IPR008271 |
| domain | Myosin Light Chain Kinase 1, Kinase domain | 693 - 951 | IPR015725 |
| binding_site | Protein kinase, ATP binding site | 702 - 725 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47633 | IMMUNOGLOBULIN |
| PANTHER Subfamily | PTHR47633:SF1 | MYOSIN LIGHT CHAIN KINASE, SMOOTH MUSCLE |
| PANTHER Protein Class | ||
| PANTHER Pathway Category |
Inflammation mediated by chemokine and cytokine signaling pathway MLCK Cytoskeletal regulation by Rho GTPase MLCK |
|
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cleavage furrow | The cleavage furrow is a plasma membrane invagination at the cell division site. The cleavage furrow begins as a shallow groove and eventually deepens to divide the cytoplasm. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| metal ion binding | Binding to a metal ion. |
| myosin light chain kinase activity | Catalysis of the reaction: ATP + myosin-light-chain = ADP + myosin-light-chain phosphate. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDFRANLQRQ | VKPKTVSEEE | RKVHSPQQVD | FRSVLAKKGT | PKTPVPEKAP | PPKPATPDFR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SVLGSKKKLP | AENGSSNAEA | LNVKATESPK | LVGNAPLSGS | LKPVANAKPA | ETLKPVANTK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PAETLKPVAN | AETLKPMGNA | KPAESSKPVG | NTKPAETLKP | VGNTKPAETL | KPVGNIKPAE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TLKPVGNIKP | AETLKPVGNT | KPTETLKPVA | NAKSAETLKP | IANTKPAETL | KPVGNAKPAE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TLKPVGNAKP | AETLKPVGNA | KPAETLKPVG | NAKPAETLKA | VANAKPAETP | KPAGKEELKK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EVQNDVNCKR | EKAGAADNEK | PPASPGTAPT | FKEKLQDVRV | AEGEKLLLQC | QVSSEPPATI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TWTLNGKTLK | TTKFVILSQE | GSLCSVSIEK | ALPEDRGLYK | CVAKNAAPEA | ECSCHVTVHD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| APASENAKAP | EMKSRRPKSS | LPPVLGTESD | ATVKKKPAPK | TPPKAATPPQ | IPQFPEDQKV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RAGERVELFG | KVAGTQPITC | TWMKFRKQIQ | DSEHIKVENS | EAGSKLTILA | ARQEHCGCYT |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LLVENKLGSR | QAQVNLTVVD | KPDPPAGTPC | ASDIRSSSLT | LSWYGSSYDG | GSAVQSYSVE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| IWDSVDKMWT | ELATCRSTSF | NVRDLLPDRE | YKFRVRAINV | YGTSEPSQES | ELTTVGEKPE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| EPKDEVEEVS | DDDEKEPEVD | YRTVTVNTEQ | KVSDFYDIEE | RLGSGKFGQV | FRLVEKKTGK |
| 730 | 740 | 750 | 760 | 770 | 780 |
| IWAGKFFKAY | SAKEKENIPA | EIGIMNCLHH | PKLVQCVDAF | EEKANIVMVL | EIVSGGELFE |
| 790 | 800 | 810 | 820 | 830 | 840 |
| RIIDEDFELT | ERECIKYMRQ | ISEGVEYIHK | QGIVHLDLKP | ENIMCVNKTG | TRIKLIDFGL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| ARRLENAGSL | KVLFGTPEFV | APEVINYEPI | SYATDMWSIG | VICYILVSGL | SPFMGDNDNE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TLANVTSATW | DFDDEAFDEI | SDDAKDFISN | LLKKDMKNRL | DCTQCLQHPW | LMKDTKNMEA |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| KKLSKDRMKK | YMARRKWQKT | GNAVRAIGRL | SSMAMISGLS | GRKSSTGSPT | SPLTAERLET |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EEDVSQAFLE | AVAEEKPHVK | PYFSKTIRDL | EVVEGSAARF | DCKIEGYPDP | EVVWFKDDQS |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| IRESRHFQID | YDEDGNCSLI | ISDVCGDDDA | KYTCKAVNSL | GEATCTAELI | VETMEEGEGE |
| GEEEEEE |