P28738
Gene name |
Kif5c (Nkhc2) |
Protein name |
Kinesin heavy chain isoform 5C |
Names |
EC 3.6.4.- , Kinesin heavy chain neuron-specific 2 , Kinesin-1 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:16574 |
EC number |
3.6.4.-: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
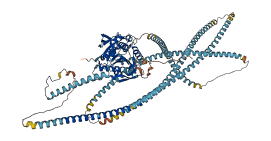
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
6-335 (Kinesin motor domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Chiba K et al. (2022) "Synergistic autoinhibition and activation mechanisms control kinesin-1 motor activity", Cell reports, 39, 110900
- Dietrich KA et al. (2008) "The kinesin-1 motor protein is regulated by a direct interaction of its head and tail", Proceedings of the National Academy of Sciences of the United States of America, 105, 8938-43
- Friedman DS et al. (1999) "Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain", Nature cell biology, 1, 293-7
- Korten T et al. (2018) "An automated in vitro motility assay for high-throughput studies of molecular motors", Lab on a chip, 18, 3196-3206
- Hannaford MR et al. (2022) "Pericentrin interacts with Kinesin-1 to drive centriole motility", The Journal of cell biology, 221,
- Sindelar CV et al. (2002) "Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy", Nature structural biology, 9, 844-8
- Kaan HY et al. (2011) "The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition", Science (New York, N.Y.), 333, 883-5
- DeBerg HA et al. (2013) "Motor domain phosphorylation modulates kinesin-1 transport", The Journal of biological chemistry, 288, 32612-32621
Autoinhibited structure

Activated structure

8 structures for P28738
43 variants for P28738
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388551290 | 52 | V>M | No | EVA | |
| rs3388549412 | 55 | P>H | No | EVA | |
| rs3388552054 | 79 | N>T | No | EVA | |
| rs3388545580 | 97 | E>K | No | EVA | |
| rs3388552434 | 102 | D>V | No | EVA | |
| rs3388551004 | 107 | G>D | No | EVA | |
| rs3388552108 | 122 | S>T | No | EVA | |
| rs3388551034 | 166 | V>A | No | EVA | |
| rs3388553298 | 187 | G>D | No | EVA | |
| rs3388551033 | 221 | E>K | No | EVA | |
| rs3391609294 | 269 | L>* | No | EVA | |
| rs3391486781 | 269 | L>M | No | EVA | |
| rs3388554374 | 319 | M>I | No | EVA | |
| rs242289747 | 335 | L>M | No | EVA | |
| rs3388550879 | 345 | K>N | No | EVA | |
| rs3388551300 | 361 | Q>H | No | EVA | |
| rs3388547569 | 381 | Q>R | No | EVA | |
| rs3388547656 | 434 | I>F | No | EVA | |
| rs3388550986 | 436 | Q>R | No | EVA | |
| rs3388555238 | 442 | E>D | No | EVA | |
| rs3391580236 | 459 | R>H | No | EVA | |
| rs3388551219 | 467 | E>* | No | EVA | |
| rs3391514900 | 489 | A>G | No | EVA | |
| rs3391425367 | 495 | V>F | No | EVA | |
| rs3388553307 | 551 | N>K | No | EVA | |
| rs3388551281 | 588 | L>P | No | EVA | |
| rs3388550872 | 609 | S>C | No | EVA | |
| rs3388552077 | 613 | D>E | No | EVA | |
| rs3388553353 | 614 | S>Y | No | EVA | |
| rs3388550960 | 628 | C>W | No | EVA | |
| rs3388547287 | 651 | Q>* | No | EVA | |
| rs3388547595 | 655 | Q>L | No | EVA | |
| rs3391542543 | 676 | M>V | No | EVA | |
| rs3388547588 | 700 | K>T | No | EVA | |
| rs3388550926 | 720 | L>R | No | EVA | |
| rs3388551322 | 723 | E>K | No | EVA | |
| rs27919511 | 765 | K>R | No | EVA | |
| rs232853282 | 768 | K>T | No | EVA | |
| rs242302146 | 773 | N>S | No | EVA | |
| rs3388547281 | 804 | F>Y | No | EVA | |
| rs244719793 | 827 | A>V | No | EVA | |
| rs3388547299 | 835 | F>L | No | EVA | |
| rs3391557877 | 852 | R>P | No | EVA |
No associated diseases with P28738
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.- | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon cytoplasm | Any cytoplasm that is part of a axon. |
| axonal growth cone | The migrating motile tip of a growing nerve cell axon. |
| ciliary rootlet | A cytoskeleton-like structure, originating from the basal body at the proximal end of a cilium, and extending proximally toward the cell nucleus. Rootlets are typically 80-100 nm in diameter and contain cross striae distributed at regular intervals of approximately 55-70 nm. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| dendrite cytoplasm | All of the contents of a dendrite, excluding the surrounding plasma membrane. |
| distal axon | That part of an axon close to and including the growth cone or the axon terminus. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| postsynaptic cytosol | The region of the cytosol consisting of all cytosol that is part of the postsynapse. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| apolipoprotein receptor binding | Binding to an apolipoprotein receptor. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
| plus-end-directed microtubule motor activity | A motor activity that generates movement along a microtubule toward the plus end, driven by ATP hydrolysis. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| anterograde axonal protein transport | The directed movement of proteins along microtubules from the cell body toward the cell periphery in nerve cell axons. |
| anterograde dendritic transport of messenger ribonucleoprotein complex | The directed movement of a messenger ribonucleoprotein complex along microtubules in nerve cell dendrites towards the postsynapse. |
| anterograde dendritic transport of neurotransmitter receptor complex | The directed movement of a neurotransmitter receptor complex along microtubules in nerve cell dendrites towards the postsynapse. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| intracellular mRNA localization | Any process in which mRNA is transported to, or maintained in, a specific location within the cell. |
| mitotic cell cycle | Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent. |
| motor neuron axon guidance | The process in which the migration of an axon growth cone of a motor neuron is directed to a specific target site in response to a combination of attractive and repulsive cues. |
| mRNA transport | The directed movement of mRNA, messenger ribonucleic acid, into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| synaptic vesicle transport | The directed movement of synaptic vesicles. |
16 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P17210 | Khc | Kinesin heavy chain | Drosophila melanogaster (Fruit fly) | EV |
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| P33175 | Kif5a | Kinesin heavy chain isoform 5A | Mus musculus (Mouse) | SS |
| Q61768 | Kif5b | Kinesin-1 heavy chain | Mus musculus (Mouse) | EV |
| Q99PW8 | Kif17 | Kinesin-like protein KIF17 | Mus musculus (Mouse) | SS |
| Q7M6Z4 | Kif27 | Kinesin-like protein KIF27 | Mus musculus (Mouse) | SS |
| Q9QXL2 | Kif21a | Kinesin-like protein KIF21A | Mus musculus (Mouse) | EV SS |
| Q9QXL1 | Kif21b | Kinesin-like protein KIF21B | Mus musculus (Mouse) | SS |
| Q9EQW7 | Kif13a | Kinesin-like protein KIF13A | Mus musculus (Mouse) | SS |
| B7ZNG0 | Kif7 | Kinesin-like protein KIF7 | Mus musculus (Mouse) | SS |
| Q6QLM7 | Kif5a | Kinesin heavy chain isoform 5A | Rattus norvegicus (Rat) | SS |
| Q2PQA9 | Kif5b | Kinesin-1 heavy chain | Rattus norvegicus (Rat) | SS |
| P34540 | unc-116 | Kinesin heavy chain | Caenorhabditis elegans | SS |
| Q9SV36 | KINUC | Kinesin-like protein KIN-UC | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MADPAECSIK | VMCRFRPLNE | AEILRGDKFI | PKFKGEETVV | IGQGKPYVFD | RVLPPNTTQE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QVYNACAKQI | VKDVLEGYNG | TIFAYGQTSS | GKTHTMEGKL | HDPQLMGIIP | RIAHDIFDHI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YSMDENLEFH | IKVSYFEIYL | DKIRDLLDVS | KTNLAVHEDK | NRVPYVKGCT | ERFVSSPEEV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MDVIDEGKAN | RHVAVTNMNE | HSSRSHSIFL | INIKQENVET | EKKLSGKLYL | VDLAGSEKVS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KTGAEGAVLD | EAKNINKSLS | ALGNVISALA | EGTKTHVPYR | DSKMTRILQD | SLGGNCRTTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VICCSPSVFN | EAETKSTLMF | GQRAKTIKNT | VSVNLELTAE | EWKKKYEKEK | EKNKALKSVL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QHLEMELNRW | RNGEAVPEDE | QISAKDQKSL | EPCDNTPIID | NITPVVDGIS | AEKEKYDEEI |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TSLYRQLDDK | DDEINQQSQL | AEKLKQQMLD | QDELLASTRR | DYEKIQEELT | RLQIENEAAK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DEVKEVLQAL | EELAVNYDQK | SQEVEDKTRA | NEQLTDELAQ | KTTTLTTTQR | ELSQLQELSN |
| 550 | 560 | 570 | 580 | 590 | 600 |
| HQKKRATEIL | NLLLKDLGEI | GGIIGTNDVK | TLADVNGVIE | EEFTMARLYI | SKMKSEVKSL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| VNRSKQLESA | QMDSNRKMNA | SERELAACQL | LISQHEAKIK | SLTDYMQNME | QKRRQLEESQ |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DSLSEELAKL | RAQEKMHEVS | FQDKEKEHLT | RLQDAEEVKK | ALEQQMESHR | EAHQKQLSRL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| RDEIEEKQRI | IDEIRDLNQK | LQLEQERLSS | DYNKLKIEDQ | EREVKLEKLL | LLNDKREQAR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EDLKGLEETV | SRELQTLHNL | RKLFVQDLTT | RVKKSVELDS | DDGGGSAAQK | QKISFLENNL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| EQLTKVHKQL | VRDNADLRCE | LPKLEKRLRA | TAERVKALES | ALKEAKENAM | RDRKRYQQEV |
| 910 | 920 | 930 | 940 | 950 | |
| DRIKEAVRAK | NMARRAHSAQ | IAKPIRPGHY | PASSPTAVHA | VRGGGGGSSN | STHYQK |