P27600
Gene name |
Gna12 (Gna-12) |
Protein name |
Guanine nucleotide-binding protein subunit alpha-12 |
Names |
G alpha-12 , G-protein subunit alpha-12 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:14673 |
EC number |
|
Protein Class |
|
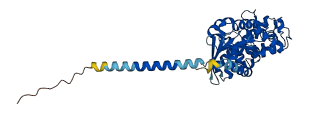
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
209-373 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Goricanec D et al. (2016) "Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding", Proceedings of the National Academy of Sciences of the United States of America, 113, E3629-38
- Coleman DE et al. (1999) "Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex", The Journal of biological chemistry, 274, 16669-72
- Lutz S et al. (2007) "Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs", Science (New York, N.Y.), 318, 1923-7
Autoinhibited structure

Activated structure

2 structures for P27600
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1ZCA | X-ray | 290 A | A/B | 48-379 | PDB |
| AF-P27600-F1 | Predicted | AlphaFoldDB |
8 variants for P27600
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3396350618 | 61 | L>V | No | EVA | |
| rs3388809646 | 114 | G>S | No | EVA | |
| rs3388798705 | 121 | E>K | No | EVA | |
| rs3388804790 | 134 | N>D | No | EVA | |
| rs3396285324 | 149 | V>G | No | EVA | |
| rs3388809619 | 173 | L>M | No | EVA | |
| rs3395926326 | 328 | Y>* | No | EVA | |
| rs3388775565 | 338 | R>K | No | EVA |
No associated diseases with P27600
No regional properties for P27600
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P27600 | |||
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| brush border membrane | The portion of the plasma membrane surrounding the brush border. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| lateral plasma membrane | The portion of the plasma membrane at the lateral side of the cell. In epithelial cells, lateral plasma membranes are on the sides of cells which lie at the interface of adjacent cells. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| D5 dopamine receptor binding | Binding to a D5 dopamine receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
| protein phosphatase 2A binding | Binding to protein phosphatase 2A. |
| protein phosphatase regulator activity | Binds to and modulates the activity of a protein phosphatase. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| cell differentiation | The cellular developmental process in which a relatively unspecialized cell, e.g. embryonic or regenerative cell, acquires specialized structural and/or functional features that characterize a specific cell. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| embryonic digit morphogenesis | The process, occurring in the embryo, by which the anatomical structures of the digit are generated and organized. A digit is one of the terminal divisions of an appendage, such as a finger or toe. |
| G protein-coupled receptor signaling pathway | The series of molecular signals initiated by a ligand binding to its receptor, in which the activated receptor promotes the exchange of GDP for GTP on the alpha-subunit of an associated heterotrimeric G-protein complex. The GTP-bound activated alpha-G-protein then dissociates from the beta- and gamma-subunits to further transmit the signal within the cell. The pathway begins with receptor-ligand interaction, and ends with regulation of a downstream cellular process. The pathway can start from the plasma membrane, Golgi or nuclear membrane. |
| in utero embryonic development | The process whose specific outcome is the progression of the embryo in the uterus over time, from formation of the zygote in the oviduct, to birth. An example of this process is found in Mus musculus. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of vascular associated smooth muscle cell migration | Any process that stops, prevents or reduces the frequency, rate or extent of vascular associated smooth muscle cell migration. |
| negative regulation of vascular associated smooth muscle cell proliferation | Any process that stops, prevents or reduces the frequency, rate or extent of vascular smooth muscle cell proliferation. |
| regulation of blood pressure | Any process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of fibroblast migration | Any process that modulates the rate, frequency or extent of fibroblast cell migration. Fibroblast cell migration is accomplished by extension and retraction of a pseudopodium. |
| regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that modulates the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| regulation of TOR signaling | Any process that modulates the frequency, rate or extent of TOR signaling. |
| response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| Rho protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rho family of proteins switching to a GTP-bound active state. |
58 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P63097 | GNAI1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P0C7Q4 | GNAT3 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04696 | GNAT2 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04695 | GNAT1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P08239 | GNAO1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P50147 | GNAI2 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P50146 | GNAI1 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P20353 | Galphai | G protein alpha i subunit | Drosophila melanogaster (Fruit fly) | SS |
| P16378 | Galphao | G protein alpha o subunit | Drosophila melanogaster (Fruit fly) | SS |
| P25157 | cta | Guanine nucleotide-binding protein subunit alpha homolog | Drosophila melanogaster (Fruit fly) | SS |
| P11488 | GNAT1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P63096 | GNAI1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | EV |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| B2RSH2 | Gnai1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| O70443 | Gnaz | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P08752 | Gnai2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P20612 | Gnat1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27601 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Mus musculus (Mouse) | SS |
| P50149 | Gnat2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| Q3V3I2 | Gnat3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| Q9DC51 | Gnai3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P21279 | Gnaq | Guanine nucleotide-binding protein G(q) subunit alpha | Mus musculus (Mouse) | PR |
| P30678 | Gna15 | Guanine nucleotide-binding protein subunit alpha-15 | Mus musculus (Mouse) | PR |
| P18872 | Gnao1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| P19627 | Gnaz | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P29348 | Gnat3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P04897 | Gnai2 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P59215 | Gnao1 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P08753 | Gnai3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q6Q7Y5 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Rattus norvegicus (Rat) | SS |
| Q63210 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| O76584 | gpa-11 | Guanine nucleotide-binding protein alpha-11 subunit | Caenorhabditis elegans | SS |
| P51875 | goa-1 | Guanine nucleotide-binding protein G | Caenorhabditis elegans | SS |
| Q18434 | odr-3 | Guanine nucleotide-binding protein alpha-17 subunit | Caenorhabditis elegans | SS |
| Q9BIG4 | gpa-10 | Guanine nucleotide-binding protein alpha-10 subunit | Caenorhabditis elegans | PR |
| P28052 | gpa-3 | Guanine nucleotide-binding protein alpha-3 subunit | Caenorhabditis elegans | SS |
| Q9XTB2 | gpa-13 | Guanine nucleotide-binding protein alpha-13 subunit | Caenorhabditis elegans | SS |
| Q93743 | gpa-6 | Guanine nucleotide-binding protein alpha-6 subunit | Caenorhabditis elegans | SS |
| Q20907 | gpa-8 | Guanine nucleotide-binding protein alpha-8 subunit | Caenorhabditis elegans | PR |
| P91907 | gpa-15 | Guanine nucleotide-binding protein alpha-15 subunit | Caenorhabditis elegans | SS |
| Q21917 | gpa-7 | Guanine nucleotide-binding protein alpha-7 subunit | Caenorhabditis elegans | SS |
| Q9BIG5 | gpa-4 | Guanine nucleotide-binding protein alpha-4 subunit | Caenorhabditis elegans | SS |
| P28051 | gpa-1 | Guanine nucleotide-binding protein alpha-1 subunit | Caenorhabditis elegans | SS |
| Q9N2V6 | gpa-16 | Guanine nucleotide-binding protein alpha-16 subunit | Caenorhabditis elegans | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGVVRTLSR | CLLPAEAGAR | ERRAGAARDA | EREARRRSRD | IDALLARERR | AVRRLVKILL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LGAGESGKST | FLKQMRIIHG | REFDQKALLE | FRDTIFDNIL | KGSRVLVDAR | DKLGIPWQHS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ENEKHGMFLM | AFENKAGLPV | EPATFQLYVP | ALSALWRDSG | IREAFSRRSE | FQLGESVKYF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LDNLDRIGQL | NYFPSKQDIL | LARKATKGIV | EHDFVIKKIP | FKMVDVGGQR | SQRQKWFQCF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DGITSILFMV | SSSEYDQVLM | EDRRTNRLVE | SMNIFETIVN | NKLFFNVSII | LFLNKMDLLV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EKVKSVSIKK | HFPDFKGDPH | RLEDVQRYLV | QCFDRKRRNR | SKPLFHHFTT | AIDTENIRFV |
| 370 | |||||
| FHAVKDTILQ | ENLKDIMLQ |