P26043
Gene name |
Rdx |
Protein name |
Radixin |
Names |
ESP10 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:19684 |
EC number |
|
Protein Class |
|
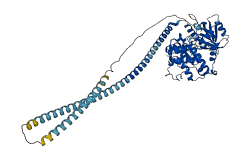
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
5-295 (FERM domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Pearson MA et al. (2000) "Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain", Cell, 101, 259-70
- Austermann J et al. (2008) "Characterization of the Ca2+ -regulated ezrin-S100P interaction and its role in tumor cell migration", The Journal of biological chemistry, 283, 29331-40
- Liu J et al. (2014) "Conserved sequence repeats of IQGAP1 mediate binding to Ezrin", Journal of proteome research, 13, 1156-66
Autoinhibited structure

Activated structure

12 structures for P26043
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1GC6 | X-ray | 290 A | A | 1-297 | PDB |
| 1GC7 | X-ray | 280 A | A | 1-297 | PDB |
| 1J19 | X-ray | 240 A | A | 1-310 | PDB |
| 2D10 | X-ray | 250 A | A/B/C/D | 1-310 | PDB |
| 2D11 | X-ray | 281 A | A/B/C/D | 1-310 | PDB |
| 2D2Q | X-ray | 280 A | A/B | 1-310 | PDB |
| 2EMS | X-ray | 290 A | A | 1-310 | PDB |
| 2EMT | X-ray | 280 A | A/B | 1-310 | PDB |
| 2YVC | X-ray | 320 A | A/B/C | 1-310 | PDB |
| 2ZPY | X-ray | 210 A | A | 1-310 | PDB |
| 3X23 | X-ray | 240 A | A | 1-310 | PDB |
| AF-P26043-F1 | Predicted | AlphaFoldDB |
19 variants for P26043
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389045772 | 30 | F>I | No | EVA | |
| rs3389041180 | 35 | K>N | No | EVA | |
| rs3388992871 | 47 | L>V | No | EVA | |
| rs3389033156 | 64 | K>E | No | EVA | |
| rs3389050566 | 199 | E>G | No | EVA | |
| rs3389041453 | 213 | G>D | No | EVA | |
| rs3389024462 | 248 | I>V | No | EVA | |
| rs3389041370 | 321 | A>T | No | EVA | |
| rs3389050564 | 328 | K>* | No | EVA | |
| rs3389050588 | 356 | E>* | No | EVA | |
| rs3389047184 | 359 | V>M | No | EVA | |
| rs3389045975 | 387 | E>D | No | EVA | |
| rs3389010905 | 389 | L>Q | No | EVA | |
| rs49859267 | 401 | S>T | No | EVA | |
| rs3389041254 | 416 | Q>H | No | EVA | |
| rs3389017983 | 460 | K>R | No | EVA | |
| rs3389050539 | 521 | N>I | No | EVA | |
| rs3389044846 | 563 | K>N | No | EVA | |
| rs3388992939 | 582 | A>S | No | EVA |
No associated diseases with P26043
10 regional properties for P26043
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 5 - 295 | IPR000299 |
| domain | Ezrin/radixin/moesin, C-terminal | 508 - 583 | IPR011259 |
| domain | FERM, N-terminal | 9 - 70 | IPR018979 |
| domain | FERM, C-terminal PH-like domain | 210 - 299 | IPR018980 |
| conserved_site | FERM conserved site | 58 - 88 | IPR019747-1 |
| conserved_site | FERM conserved site | 176 - 205 | IPR019747-2 |
| domain | FERM central domain | 90 - 206 | IPR019748 |
| domain | Band 4.1 domain | 1 - 206 | IPR019749 |
| domain | ERM family, FERM domain C-lobe | 200 - 296 | IPR041789 |
| domain | Ezrin/radixin/moesin, alpha-helical domain | 337 - 456 | IPR046810 |
Functions
15 GO annotations of cellular component
| Name | Definition |
|---|---|
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| apical part of cell | The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue. |
| cell tip | The region at the end of the longest axis of a cylindrical or elongated cell. |
| cleavage furrow | The cleavage furrow is a plasma membrane invagination at the cell division site. The cleavage furrow begins as a shallow groove and eventually deepens to divide the cytoplasm. |
| cortical actin cytoskeleton | The portion of the actin cytoskeleton, comprising filamentous actin and associated proteins, that lies just beneath the plasma membrane. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| microvillus | Thin cylindrical membrane-covered projections on the surface of an animal cell containing a core bundle of actin filaments. Present in especially large numbers on the absorptive surface of intestinal cells. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| myelin sheath | An electrically insulating fatty layer that surrounds the axons of many neurons. It is an outgrowth of glial cells |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| stereocilium | An actin-based protrusion from the apical surface of auditory and vestibular hair cells and of neuromast cells. These protrusions are supported by a bundle of cross-linked actin filaments (an actin cable), oriented such that the plus (barbed) ends are at the tip of the protrusion, capped by a tip complex which bridges to the plasma. Bundles of stereocilia act as mechanosensory organelles. |
| stereocilium base | The tapered base of the stereocilium adjacent to where it joins the hair cell body. This region contains a rootlet comprised of bundled actin filaments which spans the joint and stabilizes the stereocilium. |
| T-tubule | Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| cell adhesion molecule binding | Binding to a cell adhesion molecule. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase A binding | Binding to a protein kinase A. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| apical protein localization | Any process in which a protein is transported to, or maintained in, apical regions of the cell. |
| barbed-end actin filament capping | The binding of a protein or protein complex to the barbed (or plus) end of an actin filament, thus preventing the addition, exchange or removal of further actin subunits. |
| cellular response to thyroid hormone stimulus | A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a thyroid hormone stimulus. |
| establishment of protein localization | The directed movement of a protein to a specific location. |
| establishment of protein localization to plasma membrane | The directed movement of a protein to a specific location in a plasma membrane. |
| microvillus assembly | Formation of a microvillus, a thin cylindrical membrane-covered projection on the surface of a cell. |
| positive regulation of early endosome to late endosome transport | Any process that activates or increases the frequency, rate or extent of early endosome to late endosome transport. |
| positive regulation of G1/S transition of mitotic cell cycle | Any signalling pathway that increases or activates a cell cycle cyclin-dependent protein kinase to modulate the switch from G1 phase to S phase of the mitotic cell cycle. |
| positive regulation of protein localization to early endosome | Any process that activates or increases the frequency, rate or extent of protein localization to early endosome. |
| protein kinase A signaling | A series of reactions, mediated by the intracellular serine/threonine kinase protein kinase A, which occurs as a result of a single trigger reaction or compound. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of organelle assembly | Any process that modulates the frequency, rate or extent of organelle assembly. |
| regulation of postsynaptic neurotransmitter receptor diffusion trapping | Any process that modulates the frequency, rate or extent of postsynaptic neurotransmitter receptor diffusion trapping. |
| regulation of Rap protein signal transduction | Any process that modulates the frequency, rate or extent of Rap protein signal transduction. |
21 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P31976 | EZR | Ezrin | Bos taurus (Bovine) | PR |
| Q2HJ49 | MSN | Moesin | Bos taurus (Bovine) | PR |
| Q32LP2 | RDX | Radixin | Bos taurus (Bovine) | SS |
| Q9PU45 | RDX | Radixin | Gallus gallus (Chicken) | PR |
| Q24564 | Mer | Moesin/ezrin/radixin homolog 2 | Drosophila melanogaster (Fruit fly) | SS |
| P46150 | Moe | Moesin/ezrin/radixin homolog 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q3KP66 | INAVA | Innate immunity activator protein | Homo sapiens (Human) | PR |
| P15311 | EZR | Ezrin | Homo sapiens (Human) | EV |
| P26038 | MSN | Moesin | Homo sapiens (Human) | EV |
| P35240 | NF2 | Merlin | Homo sapiens (Human) | EV |
| P35241 | RDX | Radixin | Homo sapiens (Human) | SS |
| A2AD83 | Frmd7 | FERM domain-containing protein 7 | Mus musculus (Mouse) | PR |
| P26040 | Ezr | Ezrin | Mus musculus (Mouse) | SS |
| P26041 | Msn | Moesin | Mus musculus (Mouse) | SS |
| P46662 | Nf2 | Merlin | Mus musculus (Mouse) | SS |
| P26042 | MSN | Moesin | Sus scrofa (Pig) | PR |
| P26044 | RDX | Radixin | Sus scrofa (Pig) | SS |
| Q63648 | Nf2 | Merlin | Rattus norvegicus (Rat) | SS |
| O35763 | Msn | Moesin | Rattus norvegicus (Rat) | SS |
| P31977 | Ezr | Ezrin | Rattus norvegicus (Rat) | SS |
| Q6Q413 | nf2b | NF2, moesin-ezrin-radixin-like | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPKPINVRVT | TMDAELEFAI | QPNTTGKQLF | DQVVKTVGLR | EVWFFGLQYV | DSKGYSTWLK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LNKKVTQQDV | KKENPLQFKF | RAKFFPEDVS | EELIQEITQR | LFFLQVKEAI | LNDEIYCPPE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TAVLLASYAV | QAKYGDYNKE | IHKPGYLAND | RLLPQRVLEQ | HKLTKEQWEE | RIQNWHEEHR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GMLREDSMME | YLKIAQDLEM | YGVNYFEIKN | KKGTELWLGV | DALGLNIYEH | DDKLTPKIGF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PWSEIRNISF | NDKKFVIKPI | DKKAPDFVFY | APRLRINKRI | LALCMGNHEL | YMRRRKPDTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EVQQMKAQAR | EEKHQKQLER | AQLENEKKKR | EIAEKEKERI | EREKEELMER | LRQIEEQTVK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AQKELEEQTR | KALELEQERQ | RAKEEAERLD | RERRAAEEAK | SAIAKQAADQ | MKNQEQLAAE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LAEFTAKIAL | LEEAKKKKEE | EATEWQHKAF | AAQEDLEKTK | EELKTVMSAP | PPPPPPPVIP |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PTENEHDEQD | ENSAEASAEL | SSEGVMNHRS | EEERVTETQK | NERVKKQLQA | LSSELAQARD |
| 550 | 560 | 570 | 580 | ||
| ETKKTQNDVL | HAENVKAGRD | KYKTLRQIRQ | GNTKQRIDEF | EAM |