P26038
Gene name |
MSN |
Protein name |
Moesin |
Names |
Membrane-organizing extension spike protein |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4478 |
EC number |
|
Protein Class |
MERLIN/MOESIN/EZRIN/RADIXIN (PTHR23281) |
Descriptions
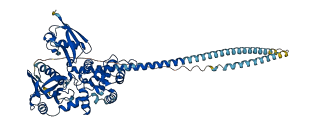
MSN is a moesin protein and belongs to the ezrin-radixin-moesin (ERM) protein family, directly involved in the cytoskeleton-membrane crosslinking. Functional N-terminal FERM domain of ERM attaches to the membrane by binding specific membrane proteins while the last 34 residues of the C-terminal tail domain bind actin filaments. The autoinhibitory domain is positions at the C-terminal tail domain of ERM, where FERM domain of ERM is bound tightly via phosphotyrosine binding (PTB), pleckstrin homology (PH), and Enabled/VASP Homology 1 (EVH1) domains, thus masking the binding sites for other molecules. ERM is activated through phosphorylation at Thr558 weakening the FERM/tail binding and, unmasks the binding sites of membrane protein and actin filaments in the presence of phospholipids.
Autoinhibitory domains (AIDs)
Target domain |
5-295 (FERM domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis |
Accessory elements
No accessory elements
References
- Pearson MA et al. (2000) "Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain", Cell, 101, 259-70
- Austermann J et al. (2008) "Characterization of the Ca2+ -regulated ezrin-S100P interaction and its role in tumor cell migration", The Journal of biological chemistry, 283, 29331-40
- Liu J et al. (2014) "Conserved sequence repeats of IQGAP1 mediate binding to Ezrin", Journal of proteome research, 13, 1156-66
Autoinhibited structure
Activated structure

10 structures for P26038
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1E5W | X-ray | 270 A | A | 2-346 | PDB |
| 1EF1 | X-ray | 190 A | PDB | ||
| 1SGH | X-ray | 350 A | A | 1-297 | PDB |
| 6TXQ | X-ray | 173 A | AAA | 1-346 | PDB |
| 6TXS | X-ray | 220 A | AAA | 1-346 | PDB |
| 8CIR | X-ray | 185 A | A/B | 1-346 | PDB |
| 8CIS | X-ray | 152 A | A | 1-346 | PDB |
| 8CIT | X-ray | 254 A | A/B/C | 1-346 | PDB |
| 8CIU | X-ray | 239 A | A | 1-346 | PDB |
| AF-P26038-F1 | Predicted | AlphaFoldDB |
198 variants for P26038
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001092797 RCV000412603 CA16042218 rs1057519074 VAR_078026 |
171 | R>W | IMD50; decreased protein abundance in T cell; loss of T cell proliferation after T cell activation; does not affect immunologic synapses formation; decreased T cell migration in response to activation by chemokines; increased T cell adhesion in response to activation by integrins Combined immunodeficiency due to moesin deficiency [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA413412570 rs1441590143 RCV001262725 |
273 | R>W | Combined immunodeficiency due to moesin deficiency [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
rs1602878106 RCV000990848 CA413414648 |
534 | D>A | Combined immunodeficiency due to moesin deficiency [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000412497 rs1057519075 CA16042219 |
553 | R>* | Combined immunodeficiency due to moesin deficiency [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA413414365 rs1311559461 |
5 | I>F | No |
ClinGen gnomAD |
|
|
rs759664613 CA10434438 |
8 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200284989 CA10434439 |
8 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1445014192 CA413414415 |
10 | T>S | No |
ClinGen TOPMed |
|
|
CA413414421 rs1334204781 |
11 | T>P | No |
ClinGen gnomAD |
|
|
CA413414466 rs1280234284 |
17 | E>A | No |
ClinGen gnomAD |
|
|
rs774220844 CA10434442 |
21 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs1482330736 CA413414497 |
22 | P>T | No |
ClinGen gnomAD |
|
|
rs1208693016 CA413414507 |
23 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs760735957 CA413414518 |
25 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs760735957 CA10434445 |
25 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA10434448 rs759935225 |
31 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA10434447 rs776735098 |
31 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1251973806 CA642290854 |
35 | K>P | No |
ClinGen gnomAD |
|
|
rs1466800155 CA413414983 |
35 | K>R | No |
ClinGen gnomAD |
|
|
rs1034323442 CA330544146 |
39 | L>S | No |
ClinGen Ensembl |
|
|
rs868321933 CA330544147 |
41 | E>K | No |
ClinGen Ensembl |
|
|
CA330544149 rs894507011 |
43 | W>C | No |
ClinGen Ensembl |
|
|
CA413415114 rs1194956967 |
54 | G>C | No |
ClinGen gnomAD |
|
|
rs182868005 CA10434460 |
57 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA413415137 rs182868005 |
57 | T>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA413415170 rs1469446074 |
62 | N>S | No |
ClinGen TOPMed |
|
|
CA10434473 rs765836962 |
69 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201686694 CA10434474 |
70 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201686694 CA413415235 |
70 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754601355 CA10434475 |
71 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA330544299 rs943287676 |
71 | R>W | No |
ClinGen gnomAD |
|
|
CA413415265 rs1460094115 |
74 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA413415272 rs1167329577 |
75 | P>L | No |
ClinGen gnomAD |
|
|
rs1223741749 CA413415267 |
75 | P>T | No |
ClinGen TOPMed |
|
|
rs1362076645 CA413415282 |
77 | L>P | No |
ClinGen TOPMed |
|
|
rs140370180 CA10434476 |
81 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10434477 rs772365732 COSM1215600 |
81 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA10434478 rs758203614 |
85 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746950857 CA10434480 |
100 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770945316 CA10434481 |
100 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1313851933 CA413415515 |
110 | I>S | No |
ClinGen gnomAD |
|
|
CA413415528 rs1315877285 |
112 | N>S | No |
ClinGen gnomAD |
|
|
CA330544301 rs982521952 |
118 | P>L | No |
ClinGen TOPMed |
|
|
CA10434485 rs775857259 |
122 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1474417388 CA413415654 |
131 | Q>H | No |
ClinGen gnomAD |
|
|
rs1602862881 CA413415674 |
134 | Y>C | No |
ClinGen Ensembl |
|
|
rs768117369 CA10434490 |
136 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs753331094 CA10434491 |
138 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413415738 rs1373451486 |
143 | K>E | No |
ClinGen gnomAD |
|
|
CA10434495 rs758172397 |
144 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs199526068 CA10434494 |
144 | S>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1221663220 CA413415748 |
145 | G>S | No |
ClinGen gnomAD |
|
|
CA10434496 rs140037207 |
148 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10434498 COSM176053 rs757131925 |
149 | G>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA413415802 rs1191782914 |
153 | L>F | No |
ClinGen gnomAD |
|
|
rs1193092131 CA413415881 |
162 | K>N | No |
ClinGen gnomAD |
|
|
CA10434514 rs763869268 |
169 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA413415946 rs1455552920 |
171 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA413415990 rs1407986048 |
177 | E>G | No |
ClinGen gnomAD |
|
|
rs1418581438 CA413415987 |
177 | E>Q | No |
ClinGen gnomAD |
|
|
CA330544385 rs886913201 COSM1124079 |
180 | R>C | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
COSM1124080 rs373340100 CA10434516 |
180 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA10434517 rs199838289 |
182 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1435551048 CA413416058 |
185 | E>G | No |
ClinGen gnomAD |
|
|
rs1306491560 CA413416081 |
189 | L>M | No |
ClinGen TOPMed |
|
|
CA10434531 rs763589764 |
193 | K>* | No |
ClinGen ExAC |
|
|
CA330544423 rs201406246 |
199 | E>D | No |
ClinGen TOPMed |
|
|
CA10434533 rs761463986 |
201 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA10434535 rs750279044 |
206 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs756098957 CA10434536 |
209 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA413416278 rs1177725877 |
217 | W>R | No |
ClinGen TOPMed |
|
|
rs753856958 CA10434538 |
220 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1473493317 CA413416297 |
220 | V>M | No |
ClinGen TOPMed |
|
|
CA413416368 rs1195702879 |
230 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs781669526 CA10434548 |
234 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1271572614 CA413416526 |
246 | R>K | No |
ClinGen Ensembl |
|
|
rs11556591 CA330544499 |
247 | N>S | No |
ClinGen Ensembl |
|
|
rs1257941102 CA413416546 |
249 | S>P | No |
ClinGen gnomAD |
|
|
CA10434551 rs773831803 |
251 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413416561 rs1450253316 |
251 | N>Y | No |
ClinGen TOPMed |
|
|
CA413416605 rs1361606523 |
257 | I>V | No |
ClinGen gnomAD |
|
|
CA330544500 rs12856307 |
264 | A>D | No |
ClinGen Ensembl |
|
|
rs760611882 CA10434555 |
265 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs139317006 CA10434554 |
265 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10434566 rs375381171 |
268 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs746416059 CA10434567 |
270 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA330544629 rs1036496677 |
270 | Y>H | No |
ClinGen TOPMed |
|
|
CA413412568 rs1194926272 |
272 | P>L | No |
ClinGen gnomAD |
|
|
rs945420266 CA330544630 |
272 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA413412571 rs867353730 |
273 | R>P | No |
ClinGen gnomAD |
|
|
CA330544631 rs867353730 |
273 | R>Q | No |
ClinGen gnomAD |
|
|
rs899329049 CA330544632 |
275 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs747717262 CA10434570 |
279 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs778459733 CA10434569 |
279 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA10434571 COSM1124083 rs771845314 |
282 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
CA330544633 rs932598480 |
284 | C>F | No |
ClinGen Ensembl |
|
|
CA413412695 rs1459799221 |
292 | M>L | No |
ClinGen TOPMed |
|
|
rs1379781650 CA413412703 |
293 | R>G | No |
ClinGen gnomAD |
|
|
rs1466860878 CA413412711 |
294 | R>H | No |
ClinGen gnomAD |
|
|
rs1167624194 CA413412718 |
295 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA413412751 rs1569468642 |
300 | I>T | No |
ClinGen Ensembl |
|
|
rs1332152716 CA413412822 |
310 | R>W | No |
ClinGen gnomAD |
|
|
rs770882351 CA10434574 |
318 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs150023347 CA10434575 |
319 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10434577 COSM1257994 rs765146391 |
320 | R>H | oesophagus [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1456206908 CA413412914 |
321 | A>T | No |
ClinGen TOPMed |
|
|
rs1602873115 CA413412926 |
322 | M>I | No |
ClinGen Ensembl |
|
|
rs1374521523 CA413412921 |
322 | M>V | No |
ClinGen gnomAD |
|
|
rs866336389 CA330544734 |
324 | E>K | No |
ClinGen Ensembl |
|
|
rs1342104235 CA413412984 |
330 | R>H | No |
ClinGen gnomAD |
|
|
rs1218915738 CA413413004 |
332 | M>I | No |
ClinGen gnomAD |
|
|
rs1159456307 CA413413029 |
336 | E>A | No |
ClinGen TOPMed |
|
|
rs777528980 CA10434590 |
336 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA413413064 rs1265457461 |
340 | I>M | No |
ClinGen gnomAD |
|
|
rs374474374 CA10434591 |
340 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs745720734 CA10434594 |
342 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA10434593 rs776474722 |
342 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs769709810 CA10434596 |
345 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1177444733 CA413413106 |
346 | E>D | No |
ClinGen TOPMed |
|
|
rs1417412293 CA413413108 |
347 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA10434598 rs200135811 COSM227664 |
349 | E>G | lung skin [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA10434597 rs775305283 |
349 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
COSM1124085 rs182327269 CA330544735 |
350 | R>S | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes |
|
CA330544736 rs927028988 |
351 | L>R | No |
ClinGen Ensembl |
|
|
rs764186464 CA10434599 |
352 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs145356792 CA10434601 |
355 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10434602 rs767686320 |
361 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA10434616 rs759526715 |
364 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10434617 rs774396765 |
369 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA10434618 rs761864712 |
370 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10434619 rs767712102 |
370 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA10434620 rs773318621 |
371 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA413413294 rs1310782775 |
372 | A>V | No |
ClinGen gnomAD |
|
|
rs992565377 CA330544756 |
373 | L>M | No |
ClinGen gnomAD |
|
|
rs1421920149 CA413413309 |
375 | L>F | No |
ClinGen TOPMed |
|
|
rs766622413 CA10434622 |
379 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10434623 rs754305867 |
381 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA10434624 COSM1124087 rs368873074 |
381 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA413413354 rs1569468998 |
382 | A>S | No |
ClinGen Ensembl |
|
|
rs1440273976 CA413413365 |
383 | Q>H | No |
ClinGen gnomAD |
|
|
rs756735394 CA10434628 |
388 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs780850640 CA330544757 |
388 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA413413410 rs1353567681 |
390 | A>D | No |
ClinGen gnomAD |
|
|
CA10434630 rs750077484 |
390 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1353567681 CA413413411 |
390 | A>V | No |
ClinGen gnomAD |
|
|
CA413413432 rs1482943021 |
393 | R>H | No |
ClinGen TOPMed |
|
|
CA10434631 rs767552861 |
394 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1461795282 CA413413443 |
395 | E>Q | No |
ClinGen gnomAD |
|
|
CA413413468 rs1448225501 |
398 | E>G | No |
ClinGen gnomAD |
|
|
CA10434637 rs748200741 |
408 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413413532 rs1196941892 |
408 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA10434636 rs748200741 |
408 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413413615 rs1453282887 |
418 | A>T | No |
ClinGen gnomAD |
|
|
CA10434653 rs748066778 |
421 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA413413655 rs1394870501 |
423 | E>D | No |
ClinGen TOPMed |
|
|
rs772096206 CA10434654 |
426 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777701828 CA10434655 |
427 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA413413717 rs1403216287 |
433 | M>I | No |
ClinGen TOPMed |
|
|
rs1412508043 CA413413720 |
434 | A>T | No |
ClinGen TOPMed |
|
|
CA10434656 COSM296775 rs371555667 |
435 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA10434657 rs771213373 |
438 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413413770 rs1240582009 |
441 | E>Q | No |
ClinGen TOPMed |
|
|
rs1177254770 CA413413789 |
444 | E>K | No |
ClinGen TOPMed |
|
|
rs777009790 CA413413814 |
447 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777009790 CA10434658 |
447 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413413848 rs1361532206 |
450 | Q>E | No |
ClinGen gnomAD |
|
|
CA330544858 rs373156244 |
451 | M>V | No |
ClinGen ESP |
|
|
CA10434669 rs764984537 |
459 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA330544859 rs772017818 |
460 | R>H | No |
ClinGen Ensembl |
|
|
rs1342406576 CA413413944 |
463 | L>V | No |
ClinGen gnomAD |
|
|
rs758263845 CA10434671 |
466 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs777771935 CA10434672 |
467 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10434674 rs757438815 |
471 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA10434675 rs781526249 |
472 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs770193594 CA10434678 |
491 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs761952339 CA10434680 |
495 | R>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1169768462 CA413414165 |
495 | R>W | No |
ClinGen gnomAD |
|
|
rs1160945080 CA413414180 |
497 | D>V | No |
ClinGen gnomAD |
|
|
CA413414190 rs1396108798 |
499 | M>V | No |
ClinGen TOPMed |
|
|
rs1403645198 CA413414242 |
506 | E>K | No |
ClinGen TOPMed |
|
|
CA413414263 rs1465848452 |
508 | R>H | No |
ClinGen gnomAD |
|
|
CA413414270 rs1171114349 |
510 | T>P | No |
ClinGen gnomAD |
|
|
rs1305168909 CA413414284 |
512 | A>T | No |
ClinGen TOPMed |
|
|
CA10434686 rs759176072 |
518 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1309870781 CA413414361 |
521 | H>Q | No |
ClinGen gnomAD |
|
|
rs144972292 CA10434698 |
524 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000788875 CA413414604 rs774679103 |
527 | S>* | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA413414602 rs1398275807 |
527 | S>A | No |
ClinGen gnomAD |
|
|
CA10434699 rs774679103 |
527 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772615370 CA10434701 |
531 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772615370 CA10434702 |
531 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10434703 rs759051413 |
537 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs753213978 CA10434705 |
540 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1321890760 CA413414719 |
544 | I>N | No |
ClinGen TOPMed |
|
|
CA413414754 rs1404562723 |
549 | M>V | No |
ClinGen TOPMed |
|
|
CA330544889 COSM1124090 rs865998710 |
550 | R>* | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA413414826 COSM3845195 rs1417887388 |
560 | R>C | breast [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA413414943 rs1488457028 |
576 | S>F | No |
ClinGen gnomAD |
1 associated diseases with P26038
[MIM: 300988]: Immunodeficiency 50 (IMD50)
A primary immunodeficiency disorder characterized by onset of recurrent bacterial or varicella zoster virus infections in early childhood, profound lymphopenia, hypogammaglobulinemia, fluctuating monocytopenia and neutropenia, and a poor immune response to vaccine antigens. {ECO:0000269|PubMed:27405666}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A primary immunodeficiency disorder characterized by onset of recurrent bacterial or varicella zoster virus infections in early childhood, profound lymphopenia, hypogammaglobulinemia, fluctuating monocytopenia and neutropenia, and a poor immune response to vaccine antigens. {ECO:0000269|PubMed:27405666}. Note=The disease is caused by variants affecting the gene represented in this entry.
10 regional properties for P26038
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 5 - 295 | IPR000299 |
| domain | Ezrin/radixin/moesin, C-terminal | 502 - 577 | IPR011259 |
| domain | FERM, N-terminal | 9 - 70 | IPR018979 |
| domain | FERM, C-terminal PH-like domain | 210 - 299 | IPR018980 |
| conserved_site | FERM conserved site | 58 - 88 | IPR019747-1 |
| conserved_site | FERM conserved site | 176 - 205 | IPR019747-2 |
| domain | FERM central domain | 91 - 206 | IPR019748 |
| domain | Band 4.1 domain | 1 - 206 | IPR019749 |
| domain | ERM family, FERM domain C-lobe | 200 - 296 | IPR041789 |
| domain | Ezrin/radixin/moesin, alpha-helical domain | 337 - 456 | IPR046810 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR23281 | MERLIN/MOESIN/EZRIN/RADIXIN |
| PANTHER Subfamily | PTHR23281:SF26 | MOESIN |
| PANTHER Protein Class |
cytoskeletal protein
actin or actin-binding cytoskeletal protein |
|
| PANTHER Pathway Category | No pathway information available | |
22 GO annotations of cellular component
| Name | Definition |
|---|---|
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| apical part of cell | The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue. |
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| basolateral plasma membrane | The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| blood microparticle | A phospholipid microvesicle that is derived from any of several cell types, such as platelets, blood cells, endothelial cells, or others, and contains membrane receptors as well as other proteins characteristic of the parental cell. Microparticles are heterogeneous in size, and are characterized as microvesicles free of nucleic acids. |
| cell periphery | The part of a cell encompassing the cell cortex, the plasma membrane, and any external encapsulating structures. |
| cell surface | The external part of the cell wall and/or plasma membrane. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| microvillus | Thin cylindrical membrane-covered projections on the surface of an animal cell containing a core bundle of actin filaments. Present in especially large numbers on the absorptive surface of intestinal cells. |
| microvillus membrane | The portion of the plasma membrane surrounding a microvillus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| pseudopodium | A temporary protrusion or retractile process of a cell, associated with flowing movements of the protoplasm, and serving for locomotion and feeding. |
| uropod | A membrane projection with related cytoskeletal components at the trailing edge of a cell in the process of migrating or being activated, found on the opposite side of the cell from the leading edge or immunological synapse, respectively. |
| vesicle | Any small, fluid-filled, spherical organelle enclosed by membrane. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| cell adhesion molecule binding | Binding to a cell adhesion molecule. |
| double-stranded RNA binding | Binding to double-stranded RNA. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| structural constituent of cytoskeleton | The action of a molecule that contributes to the structural integrity of a cytoskeletal structure. |
19 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to testosterone stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a testosterone stimulus. |
| establishment of endothelial barrier | The establishment of a barrier between endothelial cell layers, such as those in the brain, lung or intestine, to exert specific and selective control over the passage of water and solutes, thus allowing formation and maintenance of compartments that differ in fluid and solute composition. |
| establishment of epithelial cell apical/basal polarity | The specification and formation of the apicobasal polarity of an epithelial cell. |
| gland morphogenesis | The process in which the anatomical structures of a gland are generated and organized. |
| immunological synapse formation | The formation of an area of close contact between a lymphocyte (T-, B-, or natural killer cell) and a target cell through the clustering of particular signaling and adhesion molecules and their associated membrane rafts on both the lymphocyte and target cell, which facilitates activation of the lymphocyte, transfer of membrane from the target cell to the lymphocyte, and in some situations killing of the target cell through release of secretory granules and/or death-pathway ligand-receptor interaction. |
| leukocyte cell-cell adhesion | The attachment of a leukocyte to another cell via adhesion molecules. |
| leukocyte migration | The movement of a leukocyte within or between different tissues and organs of the body. |
| membrane to membrane docking | The initial attachment of a membrane to a target membrane, mediated by proteins protruding from the two membranes. Docking requires only that the membranes come close enough for the proteins to interact and adhere. |
| positive regulation of early endosome to late endosome transport | Any process that activates or increases the frequency, rate or extent of early endosome to late endosome transport. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of podosome assembly | Any process that activates or increases the rate or extent of podosome assembly. |
| positive regulation of protein localization to early endosome | Any process that activates or increases the frequency, rate or extent of protein localization to early endosome. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of cell size | Any process that modulates the size of a cell. |
| regulation of lymphocyte migration | Any process that modulates the frequency, rate or extent of lymphocyte migration. |
| regulation of organelle assembly | Any process that modulates the frequency, rate or extent of organelle assembly. |
| T cell aggregation | The adhesion of one T cell to one or more other T cells via adhesion molecules. |
| T cell migration | The movement of a T cell within or between different tissues and organs of the body. |
| T cell proliferation | The expansion of a T cell population by cell division. Follows T cell activation. |
21 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P31976 | EZR | Ezrin | Bos taurus (Bovine) | PR |
| Q2HJ49 | MSN | Moesin | Bos taurus (Bovine) | PR |
| Q32LP2 | RDX | Radixin | Bos taurus (Bovine) | SS |
| Q9PU45 | RDX | Radixin | Gallus gallus (Chicken) | PR |
| Q24564 | Mer | Moesin/ezrin/radixin homolog 2 | Drosophila melanogaster (Fruit fly) | SS |
| P46150 | Moe | Moesin/ezrin/radixin homolog 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q3KP66 | INAVA | Innate immunity activator protein | Homo sapiens (Human) | PR |
| P15311 | EZR | Ezrin | Homo sapiens (Human) | EV |
| P35240 | NF2 | Merlin | Homo sapiens (Human) | EV |
| P35241 | RDX | Radixin | Homo sapiens (Human) | SS |
| A2AD83 | Frmd7 | FERM domain-containing protein 7 | Mus musculus (Mouse) | PR |
| P26043 | Rdx | Radixin | Mus musculus (Mouse) | SS |
| P46662 | Nf2 | Merlin | Mus musculus (Mouse) | SS |
| P26040 | Ezr | Ezrin | Mus musculus (Mouse) | SS |
| P26041 | Msn | Moesin | Mus musculus (Mouse) | SS |
| P26044 | RDX | Radixin | Sus scrofa (Pig) | SS |
| P26042 | MSN | Moesin | Sus scrofa (Pig) | PR |
| Q63648 | Nf2 | Merlin | Rattus norvegicus (Rat) | SS |
| P31977 | Ezr | Ezrin | Rattus norvegicus (Rat) | SS |
| O35763 | Msn | Moesin | Rattus norvegicus (Rat) | SS |
| Q6Q413 | nf2b | NF2, moesin-ezrin-radixin-like | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPKTISVRVT | TMDAELEFAI | QPNTTGKQLF | DQVVKTIGLR | EVWFFGLQYQ | DTKGFSTWLK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LNKKVTAQDV | RKESPLLFKF | RAKFYPEDVS | EELIQDITQR | LFFLQVKEGI | LNDDIYCPPE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TAVLLASYAV | QSKYGDFNKE | VHKSGYLAGD | KLLPQRVLEQ | HKLNKDQWEE | RIQVWHEEHR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GMLREDAVLE | YLKIAQDLEM | YGVNYFSIKN | KKGSELWLGV | DALGLNIYEQ | NDRLTPKIGF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PWSEIRNISF | NDKKFVIKPI | DKKAPDFVFY | APRLRINKRI | LALCMGNHEL | YMRRRKPDTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EVQQMKAQAR | EEKHQKQMER | AMLENEKKKR | EMAEKEKEKI | EREKEELMER | LKQIEEQTKK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AQQELEEQTR | RALELEQERK | RAQSEAEKLA | KERQEAEEAK | EALLQASRDQ | KKTQEQLALE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MAELTARISQ | LEMARQKKES | EAVEWQQKAQ | MVQEDLEKTR | AELKTAMSTP | HVAEPAENEQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DEQDENGAEA | SADLRADAMA | KDRSEEERTT | EAEKNERVQK | HLKALTSELA | NARDESKKTA |
| 550 | 560 | 570 | |||
| NDMIHAENMR | LGRDKYKTLR | QIRQGNTKQR | IDEFESM |