P25723
Gene name |
tld (CG6868) |
Protein name |
Dorsal-ventral patterning protein tolloid |
Names |
|
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG6868 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
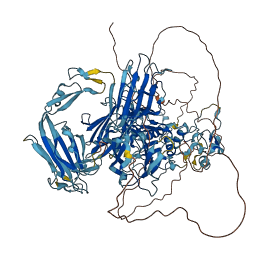
Autoinhibited structure

Activated structure

1 structures for P25723
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P25723-F1 | Predicted | AlphaFoldDB |
No variants for P25723
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P25723 | |||||
No associated diseases with P25723
16 regional properties for P25723
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| ptm | EGF-type aspartate/asparagine hydroxylation site | 606 - 617 | IPR000152-1 |
| ptm | EGF-type aspartate/asparagine hydroxylation site | 768 - 779 | IPR000152-2 |
| domain | EGF-like domain | 591 - 631 | IPR000742-1 |
| domain | EGF-like domain | 753 - 793 | IPR000742-2 |
| domain | CUB domain | 340 - 477 | IPR000859-1 |
| domain | CUB domain | 478 - 591 | IPR000859-2 |
| domain | CUB domain | 634 - 753 | IPR000859-3 |
| domain | CUB domain | 797 - 909 | IPR000859-4 |
| domain | CUB domain | 910 - 1026 | IPR000859-5 |
| domain | Peptidase M12A | 136 - 338 | IPR001506 |
| domain | EGF-like calcium-binding domain | 591 - 631 | IPR001881-1 |
| domain | EGF-like calcium-binding domain | 753 - 793 | IPR001881-2 |
| domain | Peptidase, metallopeptidase | 142 - 286 | IPR006026 |
| conserved_site | EGF-like calcium-binding, conserved site | 591 - 615 | IPR018097-1 |
| conserved_site | EGF-like calcium-binding, conserved site | 753 - 777 | IPR018097-2 |
| domain | Tolloid/BMP1 peptidase domain | 137 - 338 | IPR034036 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| collagen binding | Binding to collagen, a group of fibrous proteins of very high tensile strength that form the main component of connective tissue in animals. Collagen is highly enriched in glycine (some regions are 33% glycine) and proline, occurring predominantly as 3-hydroxyproline (about 20%). |
| metalloendopeptidase activity | Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a mechanism in which water acts as a nucleophile, one or two metal ions hold the water molecule in place, and charged amino acid side chains are ligands for the metal ions. |
| zinc ion binding | Binding to a zinc ion (Zn). |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| amnioserosa formation | Formation of the amnioserosa, an epithelium that occupies a hole in the embryonic dorsal epidermis. This occurs by the transformation of a narrow strip of cells at the dorsal midline of the blastoderm from columnar to squamous cells, accompanied by a lateral shift. |
| dorsal/ventral pattern formation | The regionalization process in which the areas along the dorsal/ventral axis are established that will lead to differences in cell differentiation. The dorsal/ventral axis is defined by a line that runs orthogonal to both the anterior/posterior and left/right axes. The dorsal end is defined by the upper or back side of an organism. The ventral end is defined by the lower or front side of an organism. |
| imaginal disc-derived wing vein morphogenesis | The process in which anatomical structures of the veins on an imaginal disc-derived wing are generated and organized. |
| maternal specification of dorsal/ventral axis, oocyte, soma encoded | Polarization of the oocyte along the dorsal-ventral axis, by a gene product encoded by somatic cells. An example of this is found in Drosophila melanogaster. |
| positive regulation of activin receptor signaling pathway | Any process that activates or increases the frequency, rate or extent of the activity of any activin receptor signaling pathway. |
| positive regulation of BMP signaling pathway | Any process that activates or increases the frequency, rate or extent of BMP signaling pathway activity. |
| protein processing | Any protein maturation process achieved by the cleavage of a peptide bond or bonds within a protein. Protein maturation is the process leading to the attainment of the full functional capacity of a protein. |
| proteolysis | The hydrolysis of proteins into smaller polypeptides and/or amino acids by cleavage of their peptide bonds. |
| R8 cell fate specification | The process in which a cell becomes capable of differentiating autonomously into an R8 cell in an environment that is neutral with respect to the developmental pathway; upon specification, the cell fate can be reversed. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9DER7 | TLL1 | Tolloid-like protein 1 | Gallus gallus (Chicken) | PR |
| P98066 | TNFAIP6 | Tumor necrosis factor-inducible gene 6 protein | Homo sapiens (Human) | PR |
| Q9Y6L7 | TLL2 | Tolloid-like protein 2 | Homo sapiens (Human) | PR |
| P13497 | BMP1 | Bone morphogenetic protein 1 | Homo sapiens (Human) | PR |
| O43897 | TLL1 | Tolloid-like protein 1 | Homo sapiens (Human) | PR |
| Q6HA09 | Astl | Astacin-like metalloendopeptidase | Mus musculus (Mouse) | PR |
| O08859 | Tnfaip6 | Tumor necrosis factor-inducible gene 6 protein | Mus musculus (Mouse) | PR |
| P98063 | Bmp1 | Bone morphogenetic protein 1 | Mus musculus (Mouse) | PR |
| Q9WVM6 | Tll2 | Tolloid-like protein 2 | Mus musculus (Mouse) | PR |
| Q62381 | Tll1 | Tolloid-like protein 1 | Mus musculus (Mouse) | PR |
| Q9U3S9 | nas-6 | Zinc metalloproteinase nas-6 | Caenorhabditis elegans | PR |
| P55112 | nas-4 | Zinc metalloproteinase nas-4 | Caenorhabditis elegans | PR |
| P55113 | nas-7 | Zinc metalloproteinase nas-7 | Caenorhabditis elegans | PR |
| Q18439 | nas-8 | Zinc metalloproteinase nas-8 | Caenorhabditis elegans | PR |
| Q20942 | nas-38 | Zinc metalloproteinase nas-38 | Caenorhabditis elegans | PR |
| Q21252 | nas-3 | Zinc metalloproteinase nas-3 | Caenorhabditis elegans | PR |
| P55115 | nas-15 | Zinc metalloproteinase nas-15 | Caenorhabditis elegans | PR |
| Q20176 | nas-39 | Zinc metalloproteinase nas-39 | Caenorhabditis elegans | PR |
| O57460 | tll1 | Dorsal-ventral patterning tolloid-like protein 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKGMRLMPMK | MKAKLVVLSV | GALWMMMFFL | VDYAEGRRLS | QLPESECDFD | FKEQPEDFFG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ILDSSLVPPK | EPKDDIYQLK | TTRQHSGRRR | KQSHKSQNKA | ALRLPPPFLW | TDDAVDVLQH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SHSPTLNGQP | IQRRRRAVTV | RKERTWDYGV | IPYEIDTIFS | GAHKALFKQA | MRHWENFTCI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KFVERDPNLH | ANYIYFTVKN | CGCCSFLGKN | GNGRQPISIG | RNCEKFGIII | HELGHTIGFH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HEHARGDRDK | HIVINKGNIM | RGQEYNFDVL | SPEEVDLPLL | PYDLNSIMHY | AKNSFSKSPY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LDTITPIGIP | PGTHLELGQR | KRLSRGDIVQ | ANLLYKCASC | GRTYQQNSGH | IVSPHFIYSG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NGVLSEFEGS | GDAGEDPSAE | SEFDASLTNC | EWRITATNGE | KVILHLQQLH | LMSSDDCTQD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YLEIRDGYWH | KSPLVRRICG | NVSGEVITTQ | TSRMLLNYVN | RNAAKGYRGF | KARFEVVCGG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DLKLTKDQSI | DSPNYPMDYM | PDKECVWRIT | APDNHQVALK | FQSFELEKHD | GCAYDFVEIR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DGNHSDSRLI | GRFCGDKLPP | NIKTRSNQMY | IRFVSDSSVQ | KLGFSAALML | DVDECKFTDH |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GCQHLCINTL | GSYQCGCRAG | YELQANGKTC | EDACGGVVDA | TKSNGSLYSP | SYPDVYPNSK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| QCVWEVVAPP | NHAVFLNFSH | FDLEGTRFHY | TKCNYDYLII | YSKMRDNRLK | KIGIYCGHEL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| PPVVNSEQSI | LRLEFYSDRT | VQRSGFVAKF | VIDVDECSMN | NGGCQHRCRN | TFGSYQCSCR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| NGYTLAENGH | NCTETRCKFE | ITTSYGVLQS | PNYPEDYPRN | IYCYWHFQTV | LGHRIQLTFH |
| 850 | 860 | 870 | 880 | 890 | 900 |
| DFEVESHQEC | IYDYVAIYDG | RSENSSTLGI | YCGGREPYAV | IASTNEMFMV | LATDAGLQRK |
| 910 | 920 | 930 | 940 | 950 | 960 |
| GFKATFVSEC | GGYLRATNHS | QTFYSHPRYG | SRPYKRNMYC | DWRIQADPES | SVKIRFLHFE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| IEYSERCDYD | YLEITEEGYS | MNTIHGRFCG | KHKPPIIISN | SDTLLLRFQT | DESNSLRGFA |
| 1030 | 1040 | 1050 | 1060 | ||
| ISFMAVDPPE | DSVGEDFDAV | TPFPGYLKSM | YSSETGSDHL | LPPSRLI |