P25490
Gene name |
YY1 (INO80S) |
Protein name |
Transcriptional repressor protein YY1 |
Names |
Delta transcription factor, INO80 complex subunit S, NF-E1, Yin and yang 1, YY-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:7528 |
EC number |
|
Protein Class |
|
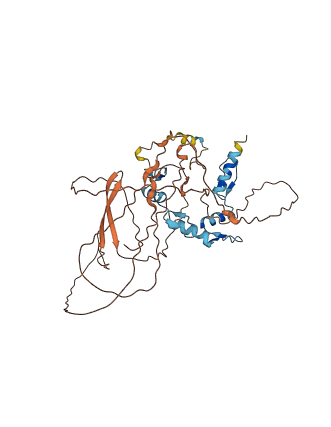
Descriptions
YY1 is a multifunctional transcription factor that exhibits positive and negative control on a large number of cellular and viral genes by binding to sites overlapping the transcription start site. This protein possesses intrinsically disordered regions (IDRs) with large negative charge, some of which involve a consecutive sequence of aspartate (D) or glutamate (E) residues, known as D/E repeats. These D/E repeats can cause autoinhibition through intramolecular electrostatic interaction with HMG boxes and modulate binding to DNA. This autoinhibited state can transition into the uninhibited complex with DNA through an electrostatically driven induced-fit process, which accelerates the target DNA search kinetics in the presence of non-functional high-affinity ligands ('decoys').
Autoinhibitory domains (AIDs)
Target domain |
296-320 (zinc-finger domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for P25490
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1UBD | X-ray | 250 A | C | 291-414 | PDB |
| 1ZNM | NMR | - | A | 352-379 | PDB |
| 4C5I | X-ray | 259 A | C | 199-228 | PDB |
| AF-P25490-F1 | Predicted | AlphaFoldDB |
196 variants for P25490
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001291657 CA390971162 rs1246486173 |
68 | A>T | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs76675246 RCV001198476 |
80 | H>missing | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs770164761 RCV002292371 CA7344016 |
144 | D>N | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs1890695996 RCV001266005 RCV001268245 |
157 | G>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1890697895 RCV001331054 |
176 | G>D | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1131692046 RCV000494728 CA390972436 |
179 | K>* | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| VAR_079202 | 179 | K>del | GADEVS [UniProt] | Yes | UniProt |
|
RCV001331052 rs1890698987 |
188 | G>S | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001266108 rs1891321014 |
319 | T>I | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_079203 | 320 | H>Y | GADEVS; unknown pathological significance [UniProt] | Yes | UniProt |
|
RCV000623107 CA390968879 rs1555370868 VAR_079204 |
339 | K>Q | Inborn genetic diseases GADEVS; unknown pathological significance [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1131692045 RCV000494726 CA390968945 |
344 | Q>* | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| VAR_079205 | 344 | Q>del | GADEVS [UniProt] | Yes | UniProt |
|
CA390969338 rs1131692163 VAR_079206 RCV000494727 |
366 | L>P | Gabriele de Vries syndrome GADEVS; reduced DNA-binding; reduced transcription regulator activity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000494721 VAR_079207 CA390969334 rs1131692044 |
366 | L>V | Gabriele de Vries syndrome GADEVS; unknown pathological significance [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001254142 rs1891340236 |
373 | H>R | Gabriele de Vries syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000494725 CA390969516 rs1131692043 VAR_065086 |
380 | D>Y | Gabriele de Vries syndrome GADEVS; reduced DNA-binding; reduced transcription regulator activity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1891340702 RCV001266764 |
387 | F>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_079208 | 393 | K>del | GADEVS; unknown pathological significance [UniProt] | Yes | UniProt |
|
rs1472368019 CA390970735 |
4 | G>R | No |
ClinGen gnomAD |
|
|
CA390970753 rs1415949028 |
6 | T>I | No |
ClinGen gnomAD |
|
|
CA390970764 rs1458971641 |
8 | Y>C | No |
ClinGen gnomAD |
|
|
CA390970761 rs1389479798 |
8 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs552013894 CA7343911 |
9 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1286455232 CA390970776 |
10 | A>S | No |
ClinGen gnomAD |
|
|
CA390970782 rs1271244582 |
11 | T>S | No |
ClinGen TOPMed |
|
|
rs1329282810 CA390970804 |
14 | S>L | No |
ClinGen gnomAD |
|
|
rs890220415 CA266822582 |
16 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA390970848 rs1210643562 |
20 | I>M | No |
ClinGen gnomAD |
|
|
rs1471534078 CA390970861 |
22 | E>D | No |
ClinGen gnomAD |
|
|
CA390970882 rs201492732 |
25 | E>D | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
|
CA390970885 rs1171014537 |
26 | I>V | No |
ClinGen gnomAD |
|
|
CA266822588 rs112723009 |
27 | E>G | No |
ClinGen Ensembl |
|
|
rs1367623299 CA390970909 |
29 | E>V | No |
ClinGen TOPMed |
|
|
CA7343917 rs777497914 |
31 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA390970930 rs770757387 |
33 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7343919 rs770757387 |
33 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776452688 CA390970947 |
35 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776452688 CA7343920 |
35 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7343923 rs774374231 |
40 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390970985 rs1595309412 |
41 | V>G | No |
ClinGen Ensembl |
|
|
rs1280409322 CA390970989 |
42 | G>D | No |
ClinGen gnomAD |
|
|
CA390971010 rs1204309662 |
45 | E>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 46 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs761956344 CA7343929 |
46 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1252510326 CA390971016 |
46 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA7343931 rs142619613 RCV000889047 |
47 | E>D | No |
ClinGen ClinVar 1000Genomes ESP TOPMed dbSNP |
|
|
CA7343930 rs767575930 |
47 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1478451548 CA390971036 |
48 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA390971030 rs1377144885 |
48 | D>N | No |
ClinGen gnomAD |
|
|
rs1388676369 CA390971043 |
49 | D>E | No |
ClinGen gnomAD |
|
|
CA266822673 rs867447858 |
49 | D>G | No |
ClinGen Ensembl |
|
|
rs968693718 CA266822677 |
50 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1452403732 CA390971063 |
52 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
rs750643631 CA7343937 |
52 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1309162432 CA390971059 |
52 | E>K | No |
ClinGen gnomAD |
|
|
rs554380259 CA266822685 |
53 | D>E | No |
ClinGen Ensembl |
|
|
rs1320953843 CA390971086 |
56 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1320953843 CA390971085 |
56 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA7343941 rs766816538 |
57 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1189077917 CA390971100 |
58 | D>G | No |
ClinGen TOPMed |
|
|
rs1467500093 CA390971112 |
60 | G>S | No |
ClinGen gnomAD |
|
|
rs201197093 CA266822708 |
64 | G>D | No |
ClinGen 1000Genomes |
|
|
rs1275256666 CA390971141 |
65 | H>N | No |
ClinGen gnomAD |
|
|
rs376271434 CA7343947 |
65 | H>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV001227270 rs1275256666 CA390971143 |
65 | H>Y | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
rs1452296158 CA390971155 |
67 | H>D | No |
ClinGen gnomAD |
|
|
CA390971160 rs777217783 |
67 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7343950 rs746643331 |
68 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7343958 rs780916558 |
70 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs780916558 CA7343959 |
70 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs1353105136 CA390971202 |
71 | H>R | No |
ClinGen Ensembl |
|
|
CA7343960 rs769652713 |
72 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1566765700 CA390971243 |
73 | H>Q | No |
ClinGen Ensembl |
|
|
rs1237081802 CA390971261 |
74 | H>Q | No |
ClinGen TOPMed |
|
|
CA266822799 rs1038824676 |
76 | H>D | No |
ClinGen TOPMed gnomAD |
|
|
rs762854228 CA390971319 |
77 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390971361 rs1397863470 |
80 | H>P | No |
ClinGen TOPMed |
|
|
rs772186005 CA7343973 |
80 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs997054119 CA266822816 |
81 | P>Q | No |
ClinGen TOPMed |
|
|
CA7343974 rs773201959 |
82 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs760925279 CA7343975 |
84 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs766727266 CA7343976 |
84 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1465323890 CA390971460 |
88 | P>S | No |
ClinGen TOPMed |
|
|
rs765758223 CA390971474 |
89 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1160058509 CA390971496 |
91 | T>P | No |
ClinGen TOPMed |
|
| TCGA novel | 94 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390971553 rs1314257490 |
97 | V>M | No |
ClinGen gnomAD |
|
|
rs1422473858 CA390971568 |
99 | H>Y | No |
ClinGen gnomAD |
|
|
CA7343985 rs756864544 |
101 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs539908009 CA7343988 |
109 | R>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1311918952 CA390971650 |
111 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1340924267 CA390971653 |
111 | E>V | No |
ClinGen gnomAD |
|
|
rs1328626457 CA390971677 |
115 | G>D | No |
ClinGen gnomAD |
|
| TCGA novel | 117 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA266822931 rs866340291 |
118 | S>* | No |
ClinGen Ensembl |
|
|
rs1208791544 CA390971724 |
120 | G>E | No |
ClinGen gnomAD |
|
|
rs759906771 CA7343997 |
120 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1249577058 CA390971735 |
121 | L>R | No |
ClinGen gnomAD |
|
|
CA390971740 rs1180677286 |
122 | R>C | No |
ClinGen gnomAD |
|
|
CA266822950 COSM242295 rs965298843 |
123 | A>T | prostate [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA390971813 rs1346667862 |
127 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs976835161 CA266822957 |
127 | F>Y | No |
ClinGen Ensembl |
|
| TCGA novel | 128 | E>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1464236898 CA390971817 |
128 | E>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
RCV001008768 rs1595309671 |
129 | D>missing | No |
ClinVar dbSNP |
|
|
rs1452149800 CA390971833 |
129 | D>N | No |
ClinGen gnomAD |
|
|
rs1306145498 CA390971909 |
135 | V>E | No |
ClinGen gnomAD |
|
|
CA7344003 rs756772698 |
136 | P>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390971916 rs1229149356 |
136 | P>S | No |
ClinGen gnomAD |
|
|
CA7344004 rs767071328 |
137 | A>V | No |
ClinGen ExAC |
|
|
CA7344005 rs750069951 |
139 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA7344006 rs755802001 |
139 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749056291 CA7344008 |
140 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA390971964 rs749056291 |
140 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA7344012 COSM2261793 rs748178185 |
141 | G>D | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed |
|
CA7344010 rs139565994 |
141 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7344014 rs776893346 |
143 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7344017 rs775885326 |
144 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770164761 CA390972008 |
144 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA7344020 rs769149627 |
145 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs769149627 CA7344019 |
145 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
rs766981294 CA390972091 |
150 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7344023 rs749983365 |
152 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs760269210 CA7344024 |
153 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7344025 rs765975441 |
155 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA390972169 rs1226735353 |
157 | G>D | No |
ClinGen gnomAD |
|
|
rs754738960 CA7344027 |
157 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7344028 rs778839522 |
158 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs752652665 CA7344030 |
159 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390972235 rs576505331 |
163 | G>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs576505331 CA7344032 |
163 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs745977468 CA7344033 |
164 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390972267 rs1389336201 |
165 | S>L | No |
ClinGen gnomAD |
|
|
CA390972262 rs1166413085 |
165 | S>P | No |
ClinGen gnomAD |
|
| TCGA novel | 166 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390972292 rs1172486228 |
167 | S>L | No |
ClinGen gnomAD |
|
|
rs1457520050 CA390972311 |
169 | G>D | No |
ClinGen gnomAD |
|
|
CA266823071 rs928224334 |
170 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA390972325 rs928224334 |
170 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1169201571 CA390972328 |
171 | R>G | No |
ClinGen TOPMed |
|
|
CA390972333 rs1373381977 |
171 | R>L | No |
ClinGen gnomAD |
|
|
CA7344035 rs780154054 |
173 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA390972363 rs1247704599 |
174 | K>Q | No |
ClinGen TOPMed |
|
|
rs1199293045 CA390972391 |
176 | G>S | No |
ClinGen TOPMed |
|
|
CA390972412 rs1319394671 |
177 | G>D | No |
ClinGen gnomAD |
|
|
CA390972445 rs1273277003 |
179 | K>N | No |
ClinGen gnomAD |
|
|
CA390972452 rs1469487250 |
180 | S>N | No |
ClinGen gnomAD |
|
|
CA390972467 rs1317050894 |
181 | G>D | No |
ClinGen TOPMed |
|
|
CA390972480 rs1247141189 |
182 | K>R | No |
ClinGen gnomAD |
|
|
rs1555368900 RCV000657546 |
184 | S>missing | No |
ClinVar dbSNP |
|
|
CA390972539 rs1436001778 |
186 | L>F | No |
ClinGen gnomAD |
|
|
CA390972536 rs1436001778 |
186 | L>V | No |
ClinGen gnomAD |
|
|
rs1423177521 CA390972555 |
187 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
CA390972571 rs1299212066 |
188 | G>V | No |
ClinGen Ensembl |
|
|
CA7344043 rs772586727 |
191 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772586727 CA390972609 |
191 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1477254750 CA390972615 |
192 | A>T | No |
ClinGen TOPMed |
|
|
rs1566766060 CA390972621 |
192 | A>V | No |
ClinGen Ensembl |
|
|
CA390972649 rs1282942458 |
195 | G>S | No |
ClinGen gnomAD |
|
|
rs1196662677 CA390972716 |
200 | P>L | No |
ClinGen TOPMed |
|
|
CA7344045 rs760179330 |
201 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA7344046 rs371252181 |
202 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1375424134 CA390972769 |
204 | K>N | No |
ClinGen Ensembl |
|
|
rs1217595948 CA390972796 |
206 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1595309901 CA390972827 |
208 | K>R | No |
ClinGen Ensembl |
|
|
rs1220415087 CA390972853 |
210 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1235535640 CA390972887 |
213 | K>Q | No |
ClinGen gnomAD |
|
|
CA390972940 rs1188606390 |
217 | G>S | No |
ClinGen gnomAD |
|
|
CA390973054 rs1409081046 |
225 | S>P | No |
ClinGen gnomAD |
|
| rs759536629 | 231 | D>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 235 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 244 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs919458830 CA266790387 |
260 | L>I | No |
ClinGen gnomAD |
|
|
rs75348257 CA7344068 |
270 | L>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7344069 rs76852509 |
270 | L>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 278 | E>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1289426653 CA390967566 |
280 | A>V | No |
ClinGen TOPMed |
|
|
rs1555370811 RCV000498533 |
287 | I>missing | No |
ClinVar dbSNP |
|
|
rs774336349 CA7344091 |
287 | I>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 289 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 292 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390968305 rs1485741517 |
299 | P>A | No |
ClinGen gnomAD |
|
|
CA390968376 rs1311599551 |
304 | T>S | No |
ClinGen TOPMed |
|
| TCGA novel | 314 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 316 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390968710 rs1189366704 |
327 | C>S | No |
ClinGen gnomAD |
|
|
rs1184263696 CA390968861 |
337 | S>R | No |
ClinGen gnomAD |
|
| TCGA novel | 339 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 358 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 363 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 363 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_074172 COSM1717684 rs386834266 CA232225 RCV000122507 |
372 | T>R | adrenal_gland found in patients with late onset insulinomas; alters DNA-binding motif; increases transactivation activity; produces a constitutive activation of cAMP and Ca2+ signaling pathways involved in insulin secretion [Cosmic, UniProt] | No |
ClinGen cosmic curated ClinVar UniProt Ensembl dbSNP |
| TCGA novel | 377 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 379 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 411 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
1 associated diseases with P25490
[MIM: 617557]: Gabriele-de Vries syndrome (GADEVS)
An autosomal dominant neurodevelopmental disorder characterized by delayed psychomotor development and intellectual disability. Most patients have behavioral and feeding problems, movement abnormalities, mild distal skeletal anomalies, and dysmorphic facial features. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant neurodevelopmental disorder characterized by delayed psychomotor development and intellectual disability. Most patients have behavioral and feeding problems, movement abnormalities, mild distal skeletal anomalies, and dysmorphic facial features. . Note=The disease is caused by variants affecting the gene represented in this entry.
3 regional properties for P25490
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | 14-3-3 protein, conserved site | 45 - 55 | IPR023409-1 |
| conserved_site | 14-3-3 protein, conserved site | 217 - 236 | IPR023409-2 |
| domain | 14-3-3 domain | 7 - 248 | IPR023410 |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin silencing complex | Any protein complex that mediates changes in chromatin structure that result in transcriptional silencing. |
| Ino80 complex | A multisubunit protein complex that contains the Ino80p ATPase; exhibits chromatin remodeling activity. |
| nuclear matrix | The dense fibrillar network lying on the inner side of the nuclear membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| PcG protein complex | A chromatin-associated multiprotein complex containing Polycomb Group proteins. In Drosophila, Polycomb group proteins are involved in the long-term maintenance of gene repression, and PcG protein complexes associate with Polycomb group response elements (PREs) in target genes to regulate higher-order chromatin structure. |
| transcription regulator complex | A protein complex that is capable of associating with DNA by direct binding, or via other DNA-binding proteins or complexes, and regulating transcription. |
15 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by some RNA polymerase. The proximal promoter is in cis with and relatively close to the core promoter. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| DNA-binding transcription repressor activity | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| four-way junction DNA binding | Binding to a DNA segment containing four-way junctions, also known as Holliday junctions, a structure where two DNA double strands are held together by reciprocal exchange of two of the four strands, one strand each from the two original helices. |
| metal ion binding | Binding to a metal ion. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| SMAD binding | Binding to a SMAD signaling protein. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
31 GO annotations of biological process
| Name | Definition |
|---|---|
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| B cell differentiation | The process in which a precursor cell type acquires the specialized features of a B cell. A B cell is a lymphocyte of B lineage with the phenotype CD19-positive and capable of B cell mediated immunity. |
| camera-type eye morphogenesis | The process in which the anatomical structures of the eye are generated and organized. The camera-type eye is an organ of sight that receives light through an aperture and focuses it through a lens, projecting it on a photoreceptor field. |
| cellular response to interleukin-1 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| cellular response to UV | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ultraviolet radiation (UV light) stimulus. Ultraviolet radiation is electromagnetic radiation with a wavelength in the range of 10 to 380 nanometers. |
| chromatin remodeling | A dynamic process of chromatin reorganization resulting in changes to chromatin structure. These changes allow DNA metabolic processes such as transcriptional regulation, DNA recombination, DNA repair, and DNA replication. |
| DNA damage response | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| double-strand break repair via homologous recombination | The error-free repair of a double-strand break in DNA in which the broken DNA molecule is repaired using homologous sequences. A strand in the broken DNA searches for a homologous region in an intact chromosome to serve as the template for DNA synthesis. The restoration of two intact DNA molecules results in the exchange, reciprocal or nonreciprocal, of genetic material between the intact DNA molecule and the broken DNA molecule. |
| immunoglobulin heavy chain V-D-J recombination | The process in which immunoglobulin heavy chain V, D, and J gene segments are recombined within a single locus utilizing the conserved heptamer and nonomer recombination signal sequences (RSS). |
| negative regulation of cell growth involved in cardiac muscle cell development | Any process that decreases the rate, frequency, or extent of the growth of a cardiac muscle cell, where growth contributes to the progression of the cell over time from its initial formation to its mature state. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of interferon-beta production | Any process that stops, prevents, or reduces the frequency, rate, or extent of interferon-beta production. |
| negative regulation of miRNA transcription | Any process that stops, prevents or reduces the frequency, rate or extent of microRNA (miRNA) gene transcription. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of DNA repair | Any process that activates or increases the frequency, rate or extent of DNA repair. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of telomere maintenance in response to DNA damage | Any process that activates or increases the frequency, rate or extent of telomere maintenance in response to DNA damage. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of chromosome organization | Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of a chromosome. |
| regulation of DNA repair | Any process that modulates the frequency, rate or extent of DNA repair. |
| regulation of DNA replication | Any process that modulates the frequency, rate or extent of DNA replication. |
| regulation of DNA strand elongation | Any process that modulates the rate, frequency or extent of DNA strand elongation. DNA strand elongation is the DNA metabolic process in which an existing DNA strand is extended by activities including the addition of nucleotides to the 3' end of the strand. |
| regulation of embryonic development | Any process that modulates the frequency, rate or extent of embryonic development. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| response to prostaglandin F | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a prostagladin F stimulus. |
| response to UV-C | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a UV-C radiation stimulus. UV-C radiation (UV-C light) spans the wavelengths 100 to 280 nm. |
| RNA localization | A process in which RNA is transported to, or maintained in, a specific location. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| telomere maintenance | Any process that contributes to the maintenance of proper telomeric length and structure by affecting and monitoring the activity of telomeric proteins, the length of telomeric DNA and the replication and repair of the DNA. These processes includes those that shorten, lengthen, replicate and repair the telomeric DNA sequences. |
177 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q08DS3 | OSR1 | Protein odd-skipped-related 1 | Bos taurus (Bovine) | PR |
| Q2VWH6 | FEZF2 | Fez family zinc finger protein 2 | Bos taurus (Bovine) | PR |
| A6QNZ0 | ZSCAN26 | Zinc finger and SCAN domain-containing protein 26 | Bos taurus (Bovine) | PR |
| A7MBI1 | ZFP69 | Zinc finger protein 69 homolog | Bos taurus (Bovine) | PR |
| Q08705 | CTCF | Transcriptional repressor CTCF | Gallus gallus (Chicken) | PR |
| O42409 | GFI1B | Zinc finger protein Gfi-1b | Gallus gallus (Chicken) | PR |
| A2T6W2 | ZNF449 | Zinc finger protein 449 | Pan troglodytes (Chimpanzee) | PR |
| Q9U405 | grau | Transcription factor grauzone | Drosophila melanogaster (Fruit fly) | PR |
| Q7K0S9 | sug | Zinc finger protein GLIS2 homolog | Drosophila melanogaster (Fruit fly) | PR |
| P20385 | Cf2 | Chorion transcription factor Cf2 | Drosophila melanogaster (Fruit fly) | PR |
| Q86P48 | ATbp | AT-rich binding protein | Drosophila melanogaster (Fruit fly) | PR |
| P28698 | MZF1 | Myeloid zinc finger 1 | Homo sapiens (Human) | PR |
| Q9NTW7 | ZFP64 | Zinc finger protein 64 | Homo sapiens (Human) | PR |
| O14978 | ZNF263 | Zinc finger protein 263 | Homo sapiens (Human) | PR |
| O60304 | ZNF500 | Zinc finger protein 500 | Homo sapiens (Human) | PR |
| P08151 | GLI1 | Zinc finger protein GLI1 | Homo sapiens (Human) | PR |
| Q9UFB7 | ZBTB47 | Zinc finger and BTB domain-containing protein 47 | Homo sapiens (Human) | PR |
| P18146 | EGR1 | Early growth response protein 1 | Homo sapiens (Human) | PR |
| Q9Y5W3 | KLF2 | Krueppel-like factor 2 | Homo sapiens (Human) | PR |
| Q9UNY5 | ZNF232 | Zinc finger protein 232 | Homo sapiens (Human) | PR |
| Q96SZ4 | ZSCAN10 | Zinc finger and SCAN domain-containing protein 10 | Homo sapiens (Human) | PR |
| P17028 | ZNF24 | Zinc finger protein 24 | Homo sapiens (Human) | PR |
| P57682 | KLF3 | Krueppel-like factor 3 | Homo sapiens (Human) | PR |
| O43296 | ZNF264 | Zinc finger protein 264 | Homo sapiens (Human) | PR |
| P49711 | CTCF | Transcriptional repressor CTCF | Homo sapiens (Human) | PR |
| Q9NQX1 | PRDM5 | PR domain zinc finger protein 5 | Homo sapiens (Human) | PR |
| Q9HBE1 | PATZ1 | POZ-, AT hook-, and zinc finger-containing protein 1 | Homo sapiens (Human) | PR |
| Q8TAX0 | OSR1 | Protein odd-skipped-related 1 | Homo sapiens (Human) | PR |
| Q9UL58 | ZNF215 | Zinc finger protein 215 | Homo sapiens (Human) | PR |
| Q8TBJ5 | FEZF2 | Fez family zinc finger protein 2 | Homo sapiens (Human) | PR |
| Q96SR6 | ZNF382 | Zinc finger protein 382 | Homo sapiens (Human) | PR |
| Q96IT1 | ZNF496 | Zinc finger protein 496 | Homo sapiens (Human) | PR |
| Q96N95 | ZNF396 | Zinc finger protein 396 | Homo sapiens (Human) | PR |
| Q9ULJ3 | ZBTB21 | Zinc finger and BTB domain-containing protein 21 | Homo sapiens (Human) | PR |
| O75840 | KLF7 | Krueppel-like factor 7 | Homo sapiens (Human) | PR |
| Q9H9D4 | ZNF408 | Zinc finger protein 408 | Homo sapiens (Human) | PR |
| Q13127 | REST | RE1-silencing transcription factor | Homo sapiens (Human) | PR |
| Q8IZM8 | ZNF654 | Zinc finger protein 654 | Homo sapiens (Human) | PR |
| Q14526 | HIC1 | Hypermethylated in cancer 1 protein | Homo sapiens (Human) | PR |
| P17022 | ZNF18 | Zinc finger protein 18 | Homo sapiens (Human) | PR |
| Q86XF7 | ZNF575 | Zinc finger protein 575 | Homo sapiens (Human) | PR |
| Q06889 | EGR3 | Early growth response protein 3 | Homo sapiens (Human) | PR |
| Q8NAM6 | ZSCAN4 | Zinc finger and SCAN domain-containing protein 4 | Homo sapiens (Human) | PR |
| Q08ER8 | ZNF543 | Zinc finger protein 543 | Homo sapiens (Human) | PR |
| P17029 | ZKSCAN1 | Zinc finger protein with KRAB and SCAN domains 1 | Homo sapiens (Human) | PR |
| Q8N680 | ZBTB2 | Zinc finger and BTB domain-containing protein 2 | Homo sapiens (Human) | PR |
| O95625 | ZBTB11 | Zinc finger and BTB domain-containing protein 11 | Homo sapiens (Human) | PR |
| Q9NPC7 | MYNN | Myoneurin | Homo sapiens (Human) | PR |
| Q96BV0 | ZNF775 | Zinc finger protein 775 | Homo sapiens (Human) | PR |
| Q8NF99 | ZNF397 | Zinc finger protein 397 | Homo sapiens (Human) | PR |
| Q63HK3 | ZKSCAN2 | Zinc finger protein with KRAB and SCAN domains 2 | Homo sapiens (Human) | PR |
| Q5FWF6 | ZNF789 | Zinc finger protein 789 | Homo sapiens (Human) | PR |
| Q15776 | ZKSCAN8 | Zinc finger protein with KRAB and SCAN domains 8 | Homo sapiens (Human) | PR |
| Q53GI3 | ZNF394 | Zinc finger protein 394 | Homo sapiens (Human) | PR |
| O95125 | ZNF202 | Zinc finger protein 202 | Homo sapiens (Human) | PR |
| Q05516 | ZBTB16 | Zinc finger and BTB domain-containing protein 16 | Homo sapiens (Human) | PR |
| Q9H116 | GZF1 | GDNF-inducible zinc finger protein 1 | Homo sapiens (Human) | PR |
| Q8N0Y2 | ZNF444 | Zinc finger protein 444 | Homo sapiens (Human) | PR |
| Q6P9G9 | ZNF449 | Zinc finger protein 449 | Homo sapiens (Human) | PR |
| Q8IW36 | ZNF695 | Zinc finger protein 695 | Homo sapiens (Human) | PR |
| Q5VTD9 | GFI1B | Zinc finger protein Gfi-1b | Homo sapiens (Human) | PR |
| Q6PG37 | ZNF790 | Zinc finger protein 790 | Homo sapiens (Human) | PR |
| Q9NQV6 | PRDM10 | PR domain zinc finger protein 10 | Homo sapiens (Human) | PR |
| Q9Y2D9 | ZNF652 | Zinc finger protein 652 | Homo sapiens (Human) | PR |
| Q5TC79 | ZBTB37 | Zinc finger and BTB domain-containing protein 37 | Homo sapiens (Human) | PR |
| Q9Y4E5 | ZNF451 | E3 SUMO-protein ligase ZNF451 | Homo sapiens (Human) | PR |
| Q8ND82 | ZNF280C | Zinc finger protein 280C | Homo sapiens (Human) | PR |
| Q49AA0 | ZFP69 | Zinc finger protein 69 homolog | Homo sapiens (Human) | PR |
| O43298 | ZBTB43 | Zinc finger and BTB domain-containing protein 43 | Homo sapiens (Human) | PR |
| Q9Y330 | ZBTB12 | Zinc finger and BTB domain-containing protein 12 | Homo sapiens (Human) | PR |
| Q13105 | ZBTB17 | Zinc finger and BTB domain-containing protein 17 | Homo sapiens (Human) | PR |
| P51508 | ZNF81 | Zinc finger protein 81 | Homo sapiens (Human) | PR |
| Q5JNZ3 | ZNF311 | Zinc finger protein 311 | Homo sapiens (Human) | PR |
| Q9BRR0 | ZKSCAN3 | Zinc finger protein with KRAB and SCAN domains 3 | Homo sapiens (Human) | PR |
| Q969J2 | ZKSCAN4 | Zinc finger protein with KRAB and SCAN domains 4 | Homo sapiens (Human) | PR |
| P49910 | ZNF165 | Zinc finger protein 165 | Homo sapiens (Human) | PR |
| Q9Y4X4 | KLF12 | Krueppel-like factor 12 | Homo sapiens (Human) | PR |
| P10074 | ZBTB48 | Telomere zinc finger-associated protein | Homo sapiens (Human) | PR |
| P17010 | ZFX | Zinc finger X-chromosomal protein | Homo sapiens (Human) | PR |
| Q9H5H4 | ZNF768 | Zinc finger protein 768 | Homo sapiens (Human) | PR |
| Q6NSZ9 | ZSCAN25 | Zinc finger and SCAN domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q9Y2L8 | ZKSCAN5 | Zinc finger protein with KRAB and SCAN domains 5 | Homo sapiens (Human) | PR |
| Q86UZ6 | ZBTB46 | Zinc finger and BTB domain-containing protein 46 | Homo sapiens (Human) | PR |
| Q9NX65 | ZSCAN32 | Zinc finger and SCAN domain-containing protein 32 | Homo sapiens (Human) | PR |
| O14771 | ZNF213 | Zinc finger protein 213 | Homo sapiens (Human) | PR |
| Q8IWY8 | ZSCAN29 | Zinc finger and SCAN domain-containing protein 29 | Homo sapiens (Human) | PR |
| Q8NCP5 | ZBTB44 | Zinc finger and BTB domain-containing protein 44 | Homo sapiens (Human) | PR |
| P41182 | BCL6 | B-cell lymphoma 6 protein | Homo sapiens (Human) | PR |
| Q9NQX0 | PRDM6 | Putative histone-lysine N-methyltransferase PRDM6 | Homo sapiens (Human) | PR |
| Q9BU19 | ZNF692 | Zinc finger protein 692 | Homo sapiens (Human) | PR |
| Q08AG5 | ZNF844 | Zinc finger protein 844 | Homo sapiens (Human) | PR |
| Q6R2W3 | ZBED9 | SCAN domain-containing protein 3 | Homo sapiens (Human) | PR |
| P98182 | ZNF200 | Zinc finger protein 200 | Homo sapiens (Human) | PR |
| Q9UK11 | ZNF223 | Zinc finger protein 223 | Homo sapiens (Human) | PR |
| O15156 | ZBTB7B | Zinc finger and BTB domain-containing protein 7B | Homo sapiens (Human) | PR |
| Q6ZMS7 | ZNF783 | Zinc finger protein 783 | Homo sapiens (Human) | PR |
| P59923 | ZNF445 | Zinc finger protein 445 | Homo sapiens (Human) | PR |
| Q8N859 | ZNF713 | Zinc finger protein 713 | Homo sapiens (Human) | PR |
| Q99612 | KLF6 | Krueppel-like factor 6 | Homo sapiens (Human) | PR |
| Q8TD17 | ZNF398 | Zinc finger protein 398 | Homo sapiens (Human) | PR |
| P52739 | ZNF131 | Zinc finger protein 131 | Homo sapiens (Human) | PR |
| A6NGD5 | ZSCAN5C | Zinc finger and SCAN domain-containing protein 5C | Homo sapiens (Human) | PR |
| Q05215 | EGR4 | Early growth response protein 4 | Homo sapiens (Human) | PR |
| Q7Z398 | ZNF550 | Zinc finger protein 550 | Homo sapiens (Human) | PR |
| Q9Y2K1 | ZBTB1 | Zinc finger and BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q96N20 | ZNF75A | Zinc finger protein 75A | Homo sapiens (Human) | PR |
| A6NJL1 | ZSCAN5B | Zinc finger and SCAN domain-containing protein 5B | Homo sapiens (Human) | PR |
| A1YPR0 | ZBTB7C | Zinc finger and BTB domain-containing protein 7C | Homo sapiens (Human) | PR |
| Q9NWS9 | ZNF446 | Zinc finger protein 446 | Homo sapiens (Human) | PR |
| P24278 | ZBTB25 | Zinc finger and BTB domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q96N38 | ZNF714 | Zinc finger protein 714 | Homo sapiens (Human) | PR |
| Q86YH2 | ZNF280B | Zinc finger protein 280B | Homo sapiens (Human) | PR |
| O08584 | Klf6 | Krueppel-like factor 6 | Mus musculus (Mouse) | PR |
| Q61164 | Ctcf | Transcriptional repressor CTCF | Mus musculus (Mouse) | PR |
| Q810A1 | Znf18 | Zinc finger protein 18 | Mus musculus (Mouse) | PR |
| Q8BGS3 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Mus musculus (Mouse) | PR |
| P41183 | Bcl6 | B-cell lymphoma 6 protein homolog | Mus musculus (Mouse) | PR |
| Q9DAI4 | Zbtb43 | Zinc finger and BTB domain-containing protein 43 | Mus musculus (Mouse) | PR |
| O70237 | Gfi1b | Zinc finger protein Gfi-1b | Mus musculus (Mouse) | PR |
| Q99KZ6 | Znf639 | Zinc finger protein 639 | Mus musculus (Mouse) | PR |
| Q9Z1D9 | Znf394 | Zinc finger protein 394 | Mus musculus (Mouse) | PR |
| Q9CXE0 | Prdm5 | PR domain zinc finger protein 5 | Mus musculus (Mouse) | PR |
| P43300 | Egr3 | Early growth response protein 3 | Mus musculus (Mouse) | PR |
| Q9DAU9 | Znf654 | Zinc finger protein 654 | Mus musculus (Mouse) | PR |
| Q9R1Y5 | Hic1 | Hypermethylated in cancer 1 protein | Mus musculus (Mouse) | PR |
| Q8R0T2 | Znf768 | Zinc finger protein 768 | Mus musculus (Mouse) | PR |
| Q9WVG7 | Osr1 | Protein odd-skipped-related 1 | Mus musculus (Mouse) | PR |
| Q8BI73 | Znf775 | Zinc finger protein 775 | Mus musculus (Mouse) | PR |
| Q8VCZ7 | Zbtb7c | Zinc finger and BTB domain-containing protein 7C | Mus musculus (Mouse) | PR |
| Q91VN1 | Znf24 | Zinc finger protein 24 | Mus musculus (Mouse) | PR |
| Q9DB38 | Znf580 | Zinc finger protein 580 | Mus musculus (Mouse) | PR |
| A7KBS4 | Zscan4d | Zinc finger and SCAN domain containing protein 4D | Mus musculus (Mouse) | PR |
| Q91VW9 | Zkscan3 | Zinc finger protein with KRAB and SCAN domains 3 | Mus musculus (Mouse) | PR |
| P10925 | Zfy1 | Zinc finger Y-chromosomal protein 1 | Mus musculus (Mouse) | PR |
| P08046 | Egr1 | Early growth response protein 1 | Mus musculus (Mouse) | PR |
| Q3UTQ7 | Prdm10 | PR domain zinc finger protein 10 | Mus musculus (Mouse) | PR |
| Q6P3Y5 | Znf280c | Zinc finger protein 280C | Mus musculus (Mouse) | PR |
| Q9ERU3 | Znf22 | Zinc finger protein 22 | Mus musculus (Mouse) | PR |
| Q8VIG1 | Rest | RE1-silencing transcription factor | Mus musculus (Mouse) | PR |
| Q9Z1D8 | Zkscan5 | Zinc finger protein with KRAB and SCAN domains 5 | Mus musculus (Mouse) | PR |
| Q8BID6 | Zbtb46 | Zinc finger and BTB domain-containing protein 46 | Mus musculus (Mouse) | PR |
| P17012 | Zfx | Zinc finger X-chromosomal protein | Mus musculus (Mouse) | PR |
| Q9WUK6 | Zbtb18 | Zinc finger and BTB domain-containing protein 18 | Mus musculus (Mouse) | PR |
| O35738 | Klf12 | Krueppel-like factor 12 | Mus musculus (Mouse) | PR |
| B2RXC5 | Znf382 | Zinc finger protein 382 | Mus musculus (Mouse) | PR |
| O08900 | Ikzf3 | Zinc finger protein Aiolos | Mus musculus (Mouse) | PR |
| Q5DU09 | Znf652 | Zinc finger protein 652 | Mus musculus (Mouse) | PR |
| Q5RJ54 | Zscan26 | Zinc finger and SCAN domain-containing protein 26 | Mus musculus (Mouse) | PR |
| Q8BLM0 | Klf8 | Krueppel-like factor 8 | Mus musculus (Mouse) | PR |
| Q99JB0 | Klf7 | Krueppel-like factor 7 | Mus musculus (Mouse) | PR |
| Q8R0A2 | Zbtb44 | Zinc finger and BTB domain-containing protein 44 | Mus musculus (Mouse) | PR |
| P20662 | Zfy2 | Zinc finger Y-chromosomal protein 2 | Mus musculus (Mouse) | PR |
| Q80VJ6 | Zscan4c | Zinc finger and SCAN domain containing protein 4C | Mus musculus (Mouse) | PR |
| Q3URS2 | Zscan4f | Zinc finger and SCAN domain containing protein 4F | Mus musculus (Mouse) | PR |
| Q60980 | Klf3 | Krueppel-like factor 3 | Mus musculus (Mouse) | PR |
| Q8K3J5 | Znf131 | Zinc finger protein 131 | Mus musculus (Mouse) | PR |
| Q00899 | Yy1 | Transcriptional repressor protein YY1 | Mus musculus (Mouse) | PR |
| Q3TTC2 | Yy2 | Transcription factor YY2 | Mus musculus (Mouse) | PR |
| Q9Z2K3 | Znf394 | Zinc finger protein 394 | Rattus norvegicus (Rat) | PR |
| Q642B9 | Znf18 | Zinc finger protein 18 | Rattus norvegicus (Rat) | PR |
| B0K011 | Osr1 | Protein odd-skipped-related 1 | Rattus norvegicus (Rat) | PR |
| D3ZUU2 | Gzf1 | GDNF-inducible zinc finger protein 1 | Rattus norvegicus (Rat) | PR |
| B1WBU4 | Zbtb8a | Zinc finger and BTB domain-containing protein 8A | Rattus norvegicus (Rat) | PR |
| Q7TNK3 | Znf24 | Zinc finger protein 24 | Rattus norvegicus (Rat) | PR |
| O35819 | Klf6 | Krueppel-like factor 6 | Rattus norvegicus (Rat) | PR |
| Q9R1D1 | Ctcf | Transcriptional repressor CTCF | Rattus norvegicus (Rat) | PR |
| P43301 | Egr3 | Early growth response protein 3 | Rattus norvegicus (Rat) | PR |
| P08154 | Egr1 | Early growth response protein 1 | Rattus norvegicus (Rat) | PR |
| A0JPL0 | Znf382 | Zinc finger protein 382 | Rattus norvegicus (Rat) | PR |
| Q4KLI1 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Rattus norvegicus (Rat) | PR |
| A1L1J6 | Znf652 | Zinc finger protein 652 | Rattus norvegicus (Rat) | PR |
| Q9SHD0 | ZAT4 | Zinc finger protein ZAT4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0P4X6 | zbtb44 | Zinc finger and BTB domain-containing protein 44 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| A4II20 | egr1 | Early growth response protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6P882 | zbtb8a.2 | Zinc finger and BTB domain-containing protein 8A.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q567C6 | znf367 | Zinc finger protein 367 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| A7Y7X5 | znf711 | Zinc finger protein 711 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASGDTLYIA | TDGSEMPAEI | VELHEIEVET | IPVETIETTV | VGEEEEEDDD | DEDGGGGDHG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GGGGHGHAGH | HHHHHHHHHH | PPMIALQPLV | TDDPTQVHHH | QEVILVQTRE | EVVGGDDSDG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRAEDGFEDQ | ILIPVPAPAG | GDDDYIEQTL | VTVAAAGKSG | GGGSSSSGGG | RVKKGGGKKS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GKKSYLSGGA | GAAGGGGADP | GNKKWEQKQV | QIKTLEGEFS | VTMWSSDEKK | DIDHETVVEE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QIIGENSPPD | YSEYMTGKKL | PPGGIPGIDL | SDPKQLAEFA | RMKPRKIKED | DAPRTIACPH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KGCTKMFRDN | SAMRKHLHTH | GPRVHVCAEC | GKAFVESSKL | KRHQLVHTGE | KPFQCTFEGC |
| 370 | 380 | 390 | 400 | 410 | |
| GKRFSLDFNL | RTHVRIHTGD | RPYVCPFDGC | NKKFAQSTNL | KSHILTHAKA | KNNQ |